
Acyl CoA dehydrogenase
Encyclopedia
Acyl-CoA dehydrogenases are a class of enzymes that function to catalyze the initial step in each cycle of fatty acid β -oxidation in the mitochondria of cells. Their action results in the introduction of a trans
double-bond between C2 and C3 of the acyl-CoA thioester
substrate. FAD
is a required co-factor in the mechanism in order for the enzyme to bind to its appropriate substrate.
The following reaction is the oxidation of the fatty acid
by FAD
.
 Acyl-CoA dehydrogenases can be categorized into three distinct groups based on their specificity for short-, medium-, or long-chain fatty acid acyl-CoA substrates. While different dehydrogenases target fatty acids of varying chain length, all types of acyl-CoA dehydrogenases are mechanistically similar. Differences in the enzyme occur based on the location of the active site along the amino acid sequence.
Acyl-CoA dehydrogenases can be categorized into three distinct groups based on their specificity for short-, medium-, or long-chain fatty acid acyl-CoA substrates. While different dehydrogenases target fatty acids of varying chain length, all types of acyl-CoA dehydrogenases are mechanistically similar. Differences in the enzyme occur based on the location of the active site along the amino acid sequence.
Acyl-CoA dehydrogenases are an important class of enzymes in mammalian cells because of their role in metabolizing fatty acids present in ingested food materials. This enzyme's action represents the first step in fatty acid metabolism (the process of breaking long chains of fatty acids into acetyl CoA molecules). Deficiencies in these enzymes are linked to genetic disorders involving fatty acid oxidation (i.e. metabolic disorders).
It is classified as .
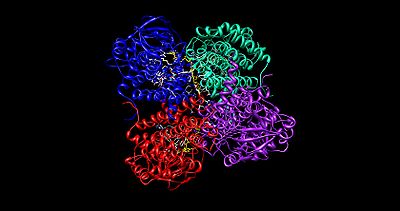 The medium chain acyl-CoA dehydrogenase is the best known structure of all acyl-CoA dehydrogenases, and is the most commonly deficient enzyme within the class that leads to metabolic disorders in animals. The molecule is a homotetramer with each subunit containing roughly 400 amino acids and one equivalent of FAD. The tetramer is classified as a “dimer of dimers” with an overall diameter of approximately 90 Å.
The medium chain acyl-CoA dehydrogenase is the best known structure of all acyl-CoA dehydrogenases, and is the most commonly deficient enzyme within the class that leads to metabolic disorders in animals. The molecule is a homotetramer with each subunit containing roughly 400 amino acids and one equivalent of FAD. The tetramer is classified as a “dimer of dimers” with an overall diameter of approximately 90 Å.
The interface between the two monomers of a single dimer of acyl-CoA dehydrogenase contains the FAD binding sites and has extensive bonding interactions. In contrast, the interface between the two dimers has few interactions. There are a total of 4 active sites within the tetramer, each of which contains a single FAD molecule and an acyl-CoA substrate. This gives a total of four FAD molecules and four acyl-CoA substrates per enzymatic molecule.
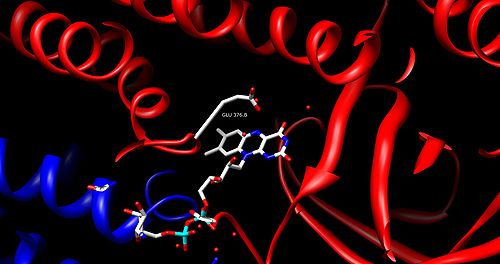
FAD is bound between the three domains of the monomer, where only the nucleotide portion is accessible. FAD binding contributes significantly to overall enzyme stability. The acyl-CoA substrate is bound completely within each monomer of the enzyme. The active site is lined with the residues F252, T255, V259, T96, T99, A100, L103, Y375, Y375, and E376. The area of interest within the substrate becomes wedged between Glu 376 and FAD, lining up the molecules into an ideal position for the reaction.
Medium-chain acyl-CoA dehydrogenases can bind to a rather broad range of chain-lengths in the acyl-CoA substrate, however studies show that its specificity tends to target octanoyl-CoA (C8-CoA).
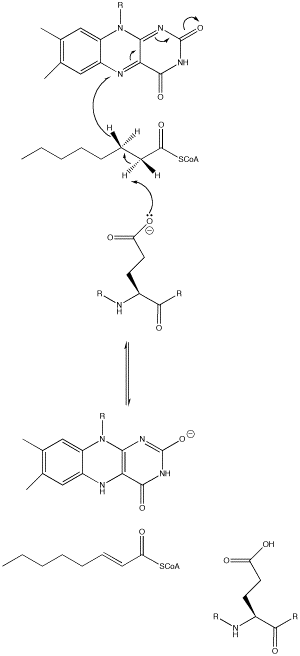 The acyl-CoA dehydrogenase mechanism proceeds through an E2 elimination. This elimination is initiated by a glutamate residue, which, while necessary for the mechanism, is not conserved.
The acyl-CoA dehydrogenase mechanism proceeds through an E2 elimination. This elimination is initiated by a glutamate residue, which, while necessary for the mechanism, is not conserved.
The residue appears in a wide range of locations within the different types of the enzyme (it is Glu 376 in medium-chain acyl-CoA dehydrogenase). The glutamate residue deprotonates the pro-R hydrogen of the alpha carbon. Hydrogen bonding of the substrate’s carbonyl oxygen to both the 2’-OH of the ribityl side-chain of FAD and to the main chain N-H of the previously mentioned glutamate residue lowers the pKa of this proton, allowing it to be readily removed by glutamate.
As the alpha carbon is being deprotonated, the pro-R hydrogen of the beta carbon leaves as a hydride to FAD. It adds to the Re face of FAD at the N-5 position, and the enzyme holds FAD in place by hydrogen bonding with the pyrimidine portion and hydrophobic interactions with the dimethylbenzene portion. The substrate has now been transformed into an alpha-beta unsaturated thioester.
As FAD picks up the hydride, the carbonyl oxygen adjacent to the N-1 nitrogen becomes negatively charged. These electrons are in resonance with the N-1 nitrogen, distributing and stabilizing the resulting negative charge. The charge is also stabilized by hydrogen bonding between the oxygen and nitrogen of interest and various residues within the enzyme.
Understanding the effects of medium-chain acyl-CoA dehydrogenase in relation to sudden infant death syndrome is of particular interest because approximately 90% of cases reveal that a mutation occurred to prevent the enzyme from properly functioning. It is reported that, every year, 1 in 20,000 infants is born with a deficiency in his/her medium-chain acyl-CoA dehydrogenases that is caused by a mutation. The mutation is recessive, and often parents of children who suffer from the deficiency can be diagnosed afterward as carriers.
In humans the most common naturally occurring mutation in the medium-chain acyl-CoA dehydrogenase is located at amino acid residue Lys-304. The altered residue occurs as a result of a single-point mutation in which the lysine side chain is replaced by a glutamate. Lys-304 typically interacts with surrounding amino acid residues by forming hydrogen bonds with Gln-342, Asp-300, and Asp-346. When a mutation causes glutamate to take the place of lysine, an additional negative charge is introduced at that site, which disrupts the normally occurring H-bonding. Such a disruption alters the folding pattern of the enzyme, ultimately compromising its stability and inhibiting its function in fatty acid oxidation. The efficiency of the mutated protein is about 10 times lower than that of the natural protein. This can lead to the symptoms of the deficiency listed above.
Trans
Trans is a Latin noun or prefix, meaning "across", "beyond" or "on the opposite side".Trans may refer to:- Science and technology :* Cis-trans isomerism, in chemistry, a form of stereoisomerism...
double-bond between C2 and C3 of the acyl-CoA thioester
Thioester
Thioesters are compounds with the functional group C-S-CO-C. They are the product of esterification between a carboxylic acid and a thiol. Thioesters are widespread in biochemistry, the best-known derivative being acetyl-CoA.-Synthesis:...
substrate. FAD
FAD
In biochemistry, flavin adenine dinucleotide is a redox cofactor involved in several important reactions in metabolism. FAD can exist in two different redox states, which it converts between by accepting or donating electrons. The molecule consists of a riboflavin moiety bound to the phosphate...
is a required co-factor in the mechanism in order for the enzyme to bind to its appropriate substrate.
The following reaction is the oxidation of the fatty acid
Fatty acid
In chemistry, especially biochemistry, a fatty acid is a carboxylic acid with a long unbranched aliphatic tail , which is either saturated or unsaturated. Most naturally occurring fatty acids have a chain of an even number of carbon atoms, from 4 to 28. Fatty acids are usually derived from...
by FAD
FAD
In biochemistry, flavin adenine dinucleotide is a redox cofactor involved in several important reactions in metabolism. FAD can exist in two different redox states, which it converts between by accepting or donating electrons. The molecule consists of a riboflavin moiety bound to the phosphate...
.

Acyl-CoA dehydrogenases are an important class of enzymes in mammalian cells because of their role in metabolizing fatty acids present in ingested food materials. This enzyme's action represents the first step in fatty acid metabolism (the process of breaking long chains of fatty acids into acetyl CoA molecules). Deficiencies in these enzymes are linked to genetic disorders involving fatty acid oxidation (i.e. metabolic disorders).
It is classified as .
Structure

The interface between the two monomers of a single dimer of acyl-CoA dehydrogenase contains the FAD binding sites and has extensive bonding interactions. In contrast, the interface between the two dimers has few interactions. There are a total of 4 active sites within the tetramer, each of which contains a single FAD molecule and an acyl-CoA substrate. This gives a total of four FAD molecules and four acyl-CoA substrates per enzymatic molecule.
FAD is bound between the three domains of the monomer, where only the nucleotide portion is accessible. FAD binding contributes significantly to overall enzyme stability. The acyl-CoA substrate is bound completely within each monomer of the enzyme. The active site is lined with the residues F252, T255, V259, T96, T99, A100, L103, Y375, Y375, and E376. The area of interest within the substrate becomes wedged between Glu 376 and FAD, lining up the molecules into an ideal position for the reaction.
Medium-chain acyl-CoA dehydrogenases can bind to a rather broad range of chain-lengths in the acyl-CoA substrate, however studies show that its specificity tends to target octanoyl-CoA (C8-CoA).
Mechanism

The residue appears in a wide range of locations within the different types of the enzyme (it is Glu 376 in medium-chain acyl-CoA dehydrogenase). The glutamate residue deprotonates the pro-R hydrogen of the alpha carbon. Hydrogen bonding of the substrate’s carbonyl oxygen to both the 2’-OH of the ribityl side-chain of FAD and to the main chain N-H of the previously mentioned glutamate residue lowers the pKa of this proton, allowing it to be readily removed by glutamate.
As the alpha carbon is being deprotonated, the pro-R hydrogen of the beta carbon leaves as a hydride to FAD. It adds to the Re face of FAD at the N-5 position, and the enzyme holds FAD in place by hydrogen bonding with the pyrimidine portion and hydrophobic interactions with the dimethylbenzene portion. The substrate has now been transformed into an alpha-beta unsaturated thioester.
As FAD picks up the hydride, the carbonyl oxygen adjacent to the N-1 nitrogen becomes negatively charged. These electrons are in resonance with the N-1 nitrogen, distributing and stabilizing the resulting negative charge. The charge is also stabilized by hydrogen bonding between the oxygen and nitrogen of interest and various residues within the enzyme.

Deficiencies of Acyl CoA dehydrogenase Linked to Metabolic Disease
Deficiencies in acyl-CoA dehydrogenases result in decreased ability to oxidize fatty acids, thereby signifying metabolic dysfunction. Medium-chain acyl-CoA dehydrogenase deficiencies MCADD are well known and characterized because they occur most commonly among acyl-CoA dehydrogenases, leading to fatty acid oxidation disorders and the potential of life-threatening metabolic diseases. Some symptoms of medium-chain acyl-CoA dehydrogenase deficiency include intolerance to fasting, hypoglycemia, and sudden infant death syndrome. These symptoms are seen as directly connected to the inability to metabolize fats. Intolerance to fasting and hypoglycemia result from the inability to gain energy and make sugar from fat stores, which is how most of humans' excess energy is stored. Also, fatty acids can begin to accumulate in the blood, lowering the blood's pKa and causing acidosis.Understanding the effects of medium-chain acyl-CoA dehydrogenase in relation to sudden infant death syndrome is of particular interest because approximately 90% of cases reveal that a mutation occurred to prevent the enzyme from properly functioning. It is reported that, every year, 1 in 20,000 infants is born with a deficiency in his/her medium-chain acyl-CoA dehydrogenases that is caused by a mutation. The mutation is recessive, and often parents of children who suffer from the deficiency can be diagnosed afterward as carriers.
In humans the most common naturally occurring mutation in the medium-chain acyl-CoA dehydrogenase is located at amino acid residue Lys-304. The altered residue occurs as a result of a single-point mutation in which the lysine side chain is replaced by a glutamate. Lys-304 typically interacts with surrounding amino acid residues by forming hydrogen bonds with Gln-342, Asp-300, and Asp-346. When a mutation causes glutamate to take the place of lysine, an additional negative charge is introduced at that site, which disrupts the normally occurring H-bonding. Such a disruption alters the folding pattern of the enzyme, ultimately compromising its stability and inhibiting its function in fatty acid oxidation. The efficiency of the mutated protein is about 10 times lower than that of the natural protein. This can lead to the symptoms of the deficiency listed above.

