CDNA library
Encyclopedia
A cDNA library is a combination of cloned cDNA (complementary DNA) fragments inserted into a collection of host cells, which together constitute some portion of the transcriptome
of the organism. cDNA is produced from fully transcribed mRNA found in the nucleus and therefore contains only the expressed genes of an organism. Similarly, tissue specific cDNA libraries can be produced. In eukaryotic cells the mature mRNA is already spliced
, hence the cDNA produced lacks introns and can be readily expressed in a bacterial cell. While information in cDNA libraries is a powerful and useful tool since gene products are easily identified, the libraries lack information about enhancers, introns, and other regulatory elements found in a genomic DNA library.
. In eukaryotes, a poly-(A) tail (consisting of a long sequence of adenine nucleotides) distinguishes mRNA from tRNA and rRNA and can therefore be used as a primer
site for reverse transcription. This has the problem that does not considers genes that do not have poly-A tail such as histone
genes.
Firstly, the mRNA is obtained and purified from the rest of the RNAs. Several methods exist for purifying RNA such as trizol
extraction and column purification. Column purification is done by using oligomeric dT nucleotide coated resins where only the mRNA having the poly-A tail will bind. The rest of the RNAs are eluted out. The mRNA is eluted by using eluting buffer and some heat to separate the mRNA strands from oligo-dT.
Once mRNA is purified, oligo-dT (a short sequence of deoxy-thymine nucleotides) is tagged as a complementary primer which binds to the poly-A tail providing a free 3'-OH end that can be extended by reverse transcriptase to create the complementary DNA strand. Now, the mRNA is removed by using a RNAse enzyme leaving a single stranded cDNA (sscDNA). This sscDNA is converted into a double stranded DNA with the help of DNA polymerase
. However, for DNA polymerase to synthesize a complementary strand a free 3'-OH end is needed. This is provided by the sscDNA itself by generating a hair pin loop at the 3' end by coiling on itself. The polymerase extends the 3'-OH end and later the loop at 3' end is opened by the scissoring action of S1 nuclease
. Restriction endonucleases and DNA ligase
are then used to clone
the sequences into bacterial plasmid
s.
The cloned bacteria are then selected, commonly through the use of antibiotic selection. Once selected, stocks of the bacteria are created which can later be grown and sequenced to compile the cDNA library.
where the additional genomic information is of less use. Also, it is useful for subsequently isolating the gene that codes for that mRNA.
Both the cDNA and the linker have blunt ends which can ligated together using a high concentration of T4 DNA ligase. Then sticky ends are produced in the cDNA molecule by cleaving the cDNA ends (which now have linkers with an incorporated site) with the appropriate endonuclease. A cloning vector (plasmid) is then also cleaved with the appropriate endonuclease. Following "sticky end" ligation of the insert into the vector the resulting recombinant DNA molecule is transferred into E. coli
host cell for cloning.
----
Transcriptome
The transcriptome is the set of all RNA molecules, including mRNA, rRNA, tRNA, and other non-coding RNA produced in one or a population of cells.-Scope:...
of the organism. cDNA is produced from fully transcribed mRNA found in the nucleus and therefore contains only the expressed genes of an organism. Similarly, tissue specific cDNA libraries can be produced. In eukaryotic cells the mature mRNA is already spliced
RNA splicing
In molecular biology and genetics, splicing is a modification of an RNA after transcription, in which introns are removed and exons are joined. This is needed for the typical eukaryotic messenger RNA before it can be used to produce a correct protein through translation...
, hence the cDNA produced lacks introns and can be readily expressed in a bacterial cell. While information in cDNA libraries is a powerful and useful tool since gene products are easily identified, the libraries lack information about enhancers, introns, and other regulatory elements found in a genomic DNA library.
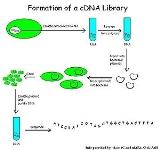
cDNA Library Construction
cDNA is created from a mature mRNA from a eukaryotic cell with the use of an enzyme known as reverse transcriptaseReverse transcriptase
In the fields of molecular biology and biochemistry, a reverse transcriptase, also known as RNA-dependent DNA polymerase, is a DNA polymerase enzyme that transcribes single-stranded RNA into single-stranded DNA. It also helps in the formation of a double helix DNA once the RNA has been reverse...
. In eukaryotes, a poly-(A) tail (consisting of a long sequence of adenine nucleotides) distinguishes mRNA from tRNA and rRNA and can therefore be used as a primer
Primer (molecular biology)
A primer is a strand of nucleic acid that serves as a starting point for DNA synthesis. They are required for DNA replication because the enzymes that catalyze this process, DNA polymerases, can only add new nucleotides to an existing strand of DNA...
site for reverse transcription. This has the problem that does not considers genes that do not have poly-A tail such as histone
Histone
In biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
genes.
mRNA extraction
Firstly, the mRNA is obtained and purified from the rest of the RNAs. Several methods exist for purifying RNA such as trizol
Trizol
TRIzol is a chemical solution used in RNA/DNA/protein extraction, by the reference paper from Piotr Chomczyński and Sacchi, N. in 1987.TRIzol is the brand name of the product from Invitrogen, and the brand name from MRC, which was founded by Chomczynski, is TRI Reagent.The correct name of the...
extraction and column purification. Column purification is done by using oligomeric dT nucleotide coated resins where only the mRNA having the poly-A tail will bind. The rest of the RNAs are eluted out. The mRNA is eluted by using eluting buffer and some heat to separate the mRNA strands from oligo-dT.
cDNA construction
Once mRNA is purified, oligo-dT (a short sequence of deoxy-thymine nucleotides) is tagged as a complementary primer which binds to the poly-A tail providing a free 3'-OH end that can be extended by reverse transcriptase to create the complementary DNA strand. Now, the mRNA is removed by using a RNAse enzyme leaving a single stranded cDNA (sscDNA). This sscDNA is converted into a double stranded DNA with the help of DNA polymerase
DNA polymerase
A DNA polymerase is an enzyme that helps catalyze in the polymerization of deoxyribonucleotides into a DNA strand. DNA polymerases are best known for their feedback role in DNA replication, in which the polymerase "reads" an intact DNA strand as a template and uses it to synthesize the new strand....
. However, for DNA polymerase to synthesize a complementary strand a free 3'-OH end is needed. This is provided by the sscDNA itself by generating a hair pin loop at the 3' end by coiling on itself. The polymerase extends the 3'-OH end and later the loop at 3' end is opened by the scissoring action of S1 nuclease
S1 nuclease
S1 nuclease is an endonuclease that is active against single-stranded DNA and RNA molecules. It is five times more active on DNA than RNA. Its reaction products are oligonucleotides or single nucleotides with 5' phosphoryl groups...
. Restriction endonucleases and DNA ligase
DNA ligase
In molecular biology, DNA ligase is a specific type of enzyme, a ligase, that repairs single-stranded discontinuities in double stranded DNA molecules, in simple words strands that have double-strand break . Purified DNA ligase is used in gene cloning to join DNA molecules together...
are then used to clone
Cloning
Cloning in biology is the process of producing similar populations of genetically identical individuals that occurs in nature when organisms such as bacteria, insects or plants reproduce asexually. Cloning in biotechnology refers to processes used to create copies of DNA fragments , cells , or...
the sequences into bacterial plasmid
Plasmid
In microbiology and genetics, a plasmid is a DNA molecule that is separate from, and can replicate independently of, the chromosomal DNA. They are double-stranded and, in many cases, circular...
s.
The cloned bacteria are then selected, commonly through the use of antibiotic selection. Once selected, stocks of the bacteria are created which can later be grown and sequenced to compile the cDNA library.
cDNA Library uses
cDNA libraries are commonly used when reproducing eukaryotic genomes, as the amount of information is reduced to remove the large numbers of non-coding regions from the library. cDNA libraries are used to express eukaryotic genes in prokaryotes. Prokaryotes do not have introns in their DNA and therefore do not possess any enzymes that can cut it out during transcription process. cDNA do not have introns and therefore can be expressed in prokaryotic cells. cDNA libraries are most useful in reverse geneticsReverse genetics
Reverse genetics is an approach to discovering the function of a gene by analyzing the phenotypic effects of specific gene sequences obtained by DNA sequencing. This investigative process proceeds in the opposite direction of so-called forward genetic screens of classical genetics...
where the additional genomic information is of less use. Also, it is useful for subsequently isolating the gene that codes for that mRNA.
cDNA Library vs. Genomic DNA Library
As previously mentioned, a cDNA library lacks the non-coding and regulatory elements found in genomic DNA. Genomic DNA libraries provide much more detailed information about the organism, but are much more resource-intensive to generate and maintain.How do we clone cDNA molecules?
cDNA molecules can be cloned by using restriction site linkers. Linkers are short, double stranded pieces of DNA (oligodeoxyribonucleotide) about 8 to 12 nucleotide pairs long that include a restriction endonuclease cleavage site e.g. BamHI.Both the cDNA and the linker have blunt ends which can ligated together using a high concentration of T4 DNA ligase. Then sticky ends are produced in the cDNA molecule by cleaving the cDNA ends (which now have linkers with an incorporated site) with the appropriate endonuclease. A cloning vector (plasmid) is then also cleaved with the appropriate endonuclease. Following "sticky end" ligation of the insert into the vector the resulting recombinant DNA molecule is transferred into E. coli
Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
host cell for cloning.
----

