
FASTA
Encyclopedia
FASTA is a DNA
and protein
sequence alignment
software package first described (as FASTP) by David J. Lipman
and William R. Pearson in 1985. Its legacy is the FASTA format
which is now ubiquitous in bioinformatics
.
sequences and DNA sequences.
The current FASTA package contains programs for protein:protein, DNA:DNA, protein:translated DNA (with frameshifts), and ordered or unordered peptide searches. Recent versions of the FASTA package include special translated search algorithms that correctly handle frameshift errors (which six-frame-translated searches do not handle very well) when comparing nucleotide to protein sequence data.
In addition to rapid heuristic search methods, the FASTA package provides SSEARCH, an implementation of the optimal Smith-Waterman algorithm.
A major focus of the package is the calculation of accurate similarity statistics, so that biologists can judge whether an alignment is likely to have occurred by chance, or whether it can be used to infer homology
. The FASTA package is available from fasta.bioch.virginia.edu.
The web-interface to submit sequences for running a search of the European Bioinformatics Institute (EBI)'s
online databases is also available using the FASTA programs.
The FASTA file format
used as input for this software is now largely used by other sequence database search tools (such as BLAST
) and sequence alignment programs (Clustal
, T-Coffee
, etc.).
The FASTA program follows a largely heuristic
method which contributes to the high speed of its execution. It initially observes the pattern of word hits, word-to-word matches of a given length, and marks potential matches before performing a more time-consuming optimized search using a Smith-Waterman type of algorithm.
The size taken for a word, given by the parameter ktup, controls the sensitivity and speed of the program. Increasing the ktup value decreases number of background hits that are found. From the word hits that are returned the program looks for segments that contain a cluster of nearby hits. It then investigates these segments for a possible match.
 ]
]
There are some differences between fastn and fastp relating to the type of sequences used but both use four steps and calculate three scores to describe and format the sequence similarity results. These are:
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
and protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
sequence alignment
Sequence alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are...
software package first described (as FASTP) by David J. Lipman
David J. Lipman
David J. Lipman is an American biologist who since 1989 has been the Director of the National Center for Biotechnology Information at the National Institutes of Health. NCBI is the home of GenBank, the U.S. node of the International Sequence Database Consortium, and PubMed, one of the most heavily...
and William R. Pearson in 1985. Its legacy is the FASTA format
FASTA format
In bioinformatics, FASTA format is a text-based format for representing either nucleotide sequences or peptide sequences, in which nucleotides or amino acids are represented using single-letter codes. The format also allows for sequence names and comments to precede the sequences...
which is now ubiquitous in bioinformatics
Bioinformatics
Bioinformatics is the application of computer science and information technology to the field of biology and medicine. Bioinformatics deals with algorithms, databases and information systems, web technologies, artificial intelligence and soft computing, information and computation theory, software...
.
History
The original FASTP program was designed for protein sequence similarity searching. FASTA added the ability to do DNA:DNA searches, translated protein:DNA searches, and also provided a more sophisticated shuffling program for evaluating statistical significance. There are several programs in this package that allow the alignment of proteinProtein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
sequences and DNA sequences.
Usage
FASTA is pronounced "fast A", and stands for "FAST-All", because it works with any alphabet, an extension of "FAST-P" (protein) and "FAST-N" (nucleotide) alignment.The current FASTA package contains programs for protein:protein, DNA:DNA, protein:translated DNA (with frameshifts), and ordered or unordered peptide searches. Recent versions of the FASTA package include special translated search algorithms that correctly handle frameshift errors (which six-frame-translated searches do not handle very well) when comparing nucleotide to protein sequence data.
In addition to rapid heuristic search methods, the FASTA package provides SSEARCH, an implementation of the optimal Smith-Waterman algorithm.
A major focus of the package is the calculation of accurate similarity statistics, so that biologists can judge whether an alignment is likely to have occurred by chance, or whether it can be used to infer homology
Homology (biology)
Homology forms the basis of organization for comparative biology. In 1843, Richard Owen defined homology as "the same organ in different animals under every variety of form and function". Organs as different as a bat's wing, a seal's flipper, a cat's paw and a human hand have a common underlying...
. The FASTA package is available from fasta.bioch.virginia.edu.
The web-interface to submit sequences for running a search of the European Bioinformatics Institute (EBI)'s
European Bioinformatics Institute
The European Bioinformatics Institute is a centre for research and services in bioinformatics, and is part of European Molecular Biology Laboratory...
online databases is also available using the FASTA programs.
The FASTA file format
FASTA format
In bioinformatics, FASTA format is a text-based format for representing either nucleotide sequences or peptide sequences, in which nucleotides or amino acids are represented using single-letter codes. The format also allows for sequence names and comments to precede the sequences...
used as input for this software is now largely used by other sequence database search tools (such as BLAST
BLAST
In bioinformatics, Basic Local Alignment Search Tool, or BLAST, is an algorithm for comparing primary biological sequence information, such as the amino-acid sequences of different proteins or the nucleotides of DNA sequences...
) and sequence alignment programs (Clustal
Clustal
Clustal is a widely used multiple sequence alignment computer program. The latest version is 2.1. There are two main variations:*ClustalW: command line interface*ClustalX: This version has a graphical user interface...
, T-Coffee
T-Coffee
T-Coffee is a multiple sequence alignment software using a progressive approach. It generates a library of pairwise alignments to guide the multiple sequence alignment...
, etc.).
Search method
FASTA takes a given nucleotide or amino-acid sequence and searches a corresponding sequence database by using local sequence alignment to find matches of similar database sequences.The FASTA program follows a largely heuristic
Heuristic
Heuristic refers to experience-based techniques for problem solving, learning, and discovery. Heuristic methods are used to speed up the process of finding a satisfactory solution, where an exhaustive search is impractical...
method which contributes to the high speed of its execution. It initially observes the pattern of word hits, word-to-word matches of a given length, and marks potential matches before performing a more time-consuming optimized search using a Smith-Waterman type of algorithm.
The size taken for a word, given by the parameter ktup, controls the sensitivity and speed of the program. Increasing the ktup value decreases number of background hits that are found. From the word hits that are returned the program looks for segments that contain a cluster of nearby hits. It then investigates these segments for a possible match.

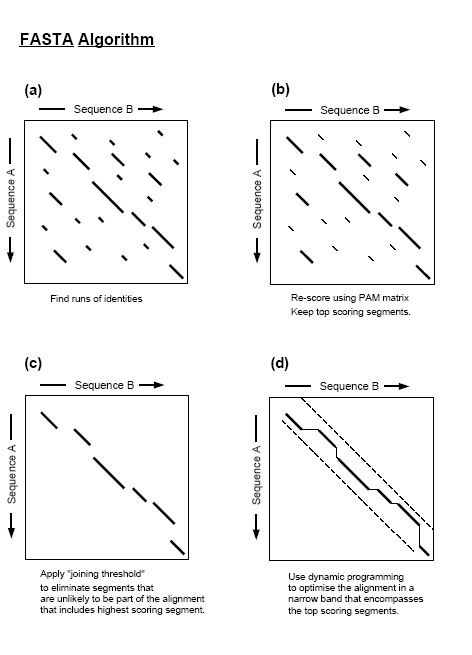
There are some differences between fastn and fastp relating to the type of sequences used but both use four steps and calculate three scores to describe and format the sequence similarity results. These are:
- Identify regions of highest density in each sequence comparison. Taking a ktup to equal 1 or 2.
- In this step all or a group of the identities between two sequences are found using a look up table. The ktup value determines how many consecutive identities are required for a match to be declared. Thus the lesser the ktup value: the more sensitive the search. ktup=2 is frequently taken by users for protein sequences and ktup=4 or 6 for nucleotide sequences. Short oligonucleotides are usually run with ktup = 1. The program then finds all similar local regions, represented as diagonals of a certain length in a dot plot, between the two sequences by counting ktup matches and penalizing for intervening mismatches. This way, local regions of highest density matches in a diagonal are isolated from background hits. For protein sequences BLOSUM50 values are used for scoring ktup matches. This ensures that groups of identities with high similarity scores contribute more to the local diagonal score than to identities with low similarity scores. Nucleotide sequences use the identity matrix for the same purpose. The best 10 local regions selected from all the diagonals put together are then saved.
- Rescan the regions taken using the scoring matrices. trimming the ends of the region to include only those contributing to the highest score.
- Rescan the 10 regions taken. This time use the relevant scoring matrix while rescoring to allow runs of identities shorter than the ktup value. Also while rescoring conservative replacements that contribute to the similarity score are taken. Though protein sequences use the BLOSUM50 matrix, scoring matrices based on the minimum number of base changes required for a specific replacement, on identities alone, or on an alternative measure of similarity such as PAM, can also be used with the program. For each of the diagonal regions rescanned this way, a subregion with the maximum score is identified. The initial scores found in step1 are used to rank the library sequences. The highest score is referred to as init1 score.
- In an alignment if several initial regions with scores greater than a CUTOFF value are found, check whether the trimmed initial regions can be joined to form an approximate alignment with gaps. Calculate a similarity score that is the sum of the joined regions penalising for each gap 20 points. This initial similarity score (initn) is used to rank the library sequences. The score of the single best initial region found in step 2 is reported (init1).
- Here the program calculates an optimal alignment of initial regions as a combination of compatible regions with maximal score. This optimal alignment of initial regions can be rapidly calculated using a dynamic programming algorithm. The resulting score initn is used to rank the library sequences.This joining process increases sensitivity but decreases selectivity. A carefully calculated cut-off value is thus used to control where this step is implemented, a value that is approximately one standard deviationStandard deviationStandard deviation is a widely used measure of variability or diversity used in statistics and probability theory. It shows how much variation or "dispersion" there is from the average...
above the average score expected from unrelated sequences in the library. A 200-residue query sequence with ktup2 uses a value 28.
- Use a banded Smith-WatermanSmith Waterman algorithmThe Smith–Waterman algorithm is a well-known algorithm for performing local sequence alignment; that is, for determining similar regions between two nucleotide or protein sequences...
algorithm to calculate an optimal score for alignment.
- This step uses a banded Smith-WatermanSmith Waterman algorithmThe Smith–Waterman algorithm is a well-known algorithm for performing local sequence alignment; that is, for determining similar regions between two nucleotide or protein sequences...
algorithm to create an optimised score (opt) for each alignment of query sequence to a database(library) sequence. It takes a band of 32 residues centered on the init1 region of step2 for calculating the optimal alignment. After all sequences are searched the program plots the initial scores of each database sequence in a histogramHistogramIn statistics, a histogram is a graphical representation showing a visual impression of the distribution of data. It is an estimate of the probability distribution of a continuous variable and was first introduced by Karl Pearson...
, and calculates the statistical significance of the "opt" score. For protein sequences, the final alignment is produced using a full Smith-WatermanSmith Waterman algorithmThe Smith–Waterman algorithm is a well-known algorithm for performing local sequence alignment; that is, for determining similar regions between two nucleotide or protein sequences...
alignment. For DNA sequences, a banded alignment is provided.
See also
- BLASTBLASTIn bioinformatics, Basic Local Alignment Search Tool, or BLAST, is an algorithm for comparing primary biological sequence information, such as the amino-acid sequences of different proteins or the nucleotides of DNA sequences...
- FASTA formatFASTA formatIn bioinformatics, FASTA format is a text-based format for representing either nucleotide sequences or peptide sequences, in which nucleotides or amino acids are represented using single-letter codes. The format also allows for sequence names and comments to precede the sequences...
- Sequence alignmentSequence alignmentIn bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are...
- Sequence alignment softwareSequence alignment softwareThis list of sequence alignment software is a compilation of software tools and web portals used in pairwise sequence alignment and multiple sequence alignment...
- Sequence profiling toolSequence profiling toolA sequence profiling tool in bioinformatics is a type of software that presents information related to a genetic sequence, gene name, or keyword input. Such tools generally take a query such as a DNA, RNA, or protein sequence or ‘keyword’ and search one or more databases for information related to...
External links
- FASTA Website
- EBI's FASTA page - EBI'sEuropean Bioinformatics InstituteThe European Bioinformatics Institute is a centre for research and services in bioinformatics, and is part of European Molecular Biology Laboratory...
page for accessing FASTA services.

