
Multiplex ligation-dependent probe amplification
Encyclopedia
Multiplex ligation-dependent probe amplification (MLPA)http://www.mlpa.com is a variation of the polymerase chain reaction
that permits multiple targets to be amplified with only a single primer
pair. Each probe consists of a two oligonucleotides which recognise adjacent target sites on the DNA
. One probe oligonucleotide contains the sequence
recognised by the forward primer, the other the sequence recognised by the reverse primer. Only when both probe oligonucleotides are hybridised to their respective targets, can they be ligated
into a complete probe. The advantage of splitting the probe into two parts is that only the ligated oligonucleotides, but not the unbound probe oligonucleotides, are amplified. If the probes were not split in this way, the primer sequences at either end would cause the probes to be amplified regardless of their hybridization to the template DNA, and the amplification product would not be dependent on the number of target sites present in the sample DNA. Each complete probe has a unique length, so that its resulting amplicons can be separated and identified by (capillary) electrophoresis
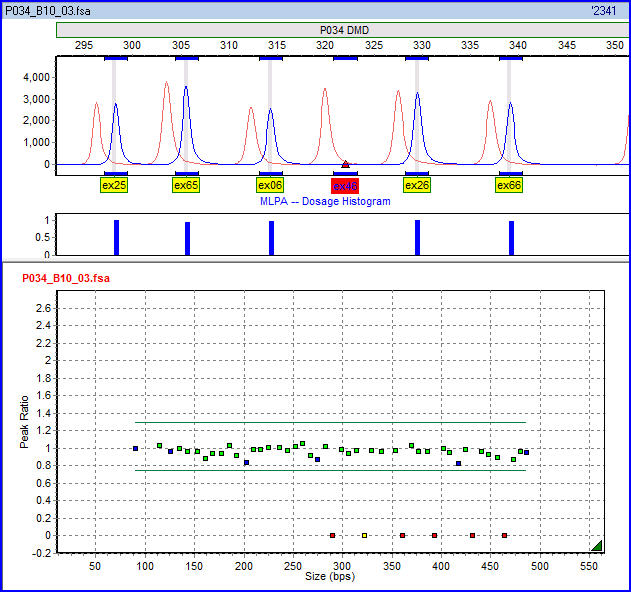
. This avoids the resolution limitations of multiplex PCR. Since the forward primer used for probe amplification is fluorescently labeled, each amplicon generates a fluorescent peak which can be detected by a capillary sequencer. Comparing the peak pattern obtained on a given sample with that obtained on various reference samples, the relative quantity of each amplicon can be determined. This ratio is a measure for the ratio in which the target sequence is present in the sample DNA.
Various techniques including DGGE (Denaturing Gradient Gel Electrophoresis), DHPLC (Denaturing High Performance Liquid Chromatography
), and SSCA (Single Strand Conformation Analysis) effectively identify SNPs and small insertions and deletions. MLPA, however, is one of the only accurate, time-efficient techniques to detect genomic deletions and insertions (one or more entire exons), which are frequent causes of cancers such as hereditary non-polyposis colorectal cancer (HNPCC), breast, and ovarian cancer. MLPA can successfully and easily determine the relative copy number of all exons within a gene simultaneously with high sensitivity.
. For example, probes may be designed to target various regions of chromosome
21 of a human cell. The signal strengths of the probes are compared with those obtained from a reference DNA sample known to have two copies of the chromosome. If an extra copy is present in the test sample, the signals are expected to be 1.5 times the intensities of the respective probes from the reference. If only one copy is present the proportion is expected to be 0.5. If the sample has two copies, the relative probe strengths are expected to be equal.
s and single nucleotide polymorphism
s, analysis of DNA methylation
, relative mRNA quantification, chromosomal characterisation of cell lines and tissue samples, detection of gene copy number,detection of duplications and deletions in human cancer
predisposition genes such as BRCA1, BRCA2, hMLH1 and hMSH2 and aneuploidy
determination. MLPA has potential application in prenatal diagnosis
both invasive and noninvasive.
Polymerase chain reaction
The polymerase chain reaction is a scientific technique in molecular biology to amplify a single or a few copies of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence....
that permits multiple targets to be amplified with only a single primer
Primer (molecular biology)
A primer is a strand of nucleic acid that serves as a starting point for DNA synthesis. They are required for DNA replication because the enzymes that catalyze this process, DNA polymerases, can only add new nucleotides to an existing strand of DNA...
pair. Each probe consists of a two oligonucleotides which recognise adjacent target sites on the DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
. One probe oligonucleotide contains the sequence
Sequence (biology)
A sequence in biology is the one-dimensional ordering of monomers, covalently linked within in a biopolymer; it is also referred to as the primary structure of the biological macromolecule.-See also:* Protein sequence* DNA sequence...
recognised by the forward primer, the other the sequence recognised by the reverse primer. Only when both probe oligonucleotides are hybridised to their respective targets, can they be ligated
Ligase
In biochemistry, ligase is an enzyme that can catalyse the joining of two large molecules by forming a new chemical bond, usually with accompanying hydrolysis of a small chemical group dependent to one of the larger molecules...
into a complete probe. The advantage of splitting the probe into two parts is that only the ligated oligonucleotides, but not the unbound probe oligonucleotides, are amplified. If the probes were not split in this way, the primer sequences at either end would cause the probes to be amplified regardless of their hybridization to the template DNA, and the amplification product would not be dependent on the number of target sites present in the sample DNA. Each complete probe has a unique length, so that its resulting amplicons can be separated and identified by (capillary) electrophoresis
Gel electrophoresis
Gel electrophoresis is a method used in clinical chemistry to separate proteins by charge and or size and in biochemistry and molecular biology to separate a mixed population of DNA and RNA fragments by length, to estimate the size of DNA and RNA fragments or to separate proteins by charge...
. This avoids the resolution limitations of multiplex PCR. Since the forward primer used for probe amplification is fluorescently labeled, each amplicon generates a fluorescent peak which can be detected by a capillary sequencer. Comparing the peak pattern obtained on a given sample with that obtained on various reference samples, the relative quantity of each amplicon can be determined. This ratio is a measure for the ratio in which the target sequence is present in the sample DNA.

Various techniques including DGGE (Denaturing Gradient Gel Electrophoresis), DHPLC (Denaturing High Performance Liquid Chromatography
SNP genotyping
SNP genotyping is the measurement of genetic variations of single nucleotide polymorphisms between members of a species. It is a form of genotyping, which is the measurement of more general genetic variation. SNPs are one of the most common types of genetic variation...
), and SSCA (Single Strand Conformation Analysis) effectively identify SNPs and small insertions and deletions. MLPA, however, is one of the only accurate, time-efficient techniques to detect genomic deletions and insertions (one or more entire exons), which are frequent causes of cancers such as hereditary non-polyposis colorectal cancer (HNPCC), breast, and ovarian cancer. MLPA can successfully and easily determine the relative copy number of all exons within a gene simultaneously with high sensitivity.
Relative ploidy
An important use of MLPA is to determine relative ploidyPloidy
Ploidy is the number of sets of chromosomes in a biological cell.Human sex cells have one complete set of chromosomes from the male or female parent. Sex cells, also called gametes, combine to produce somatic cells. Somatic cells, therefore, have twice as many chromosomes. The haploid number is...
. For example, probes may be designed to target various regions of chromosome
Chromosome
A chromosome is an organized structure of DNA and protein found in cells. It is a single piece of coiled DNA containing many genes, regulatory elements and other nucleotide sequences. Chromosomes also contain DNA-bound proteins, which serve to package the DNA and control its functions.Chromosomes...
21 of a human cell. The signal strengths of the probes are compared with those obtained from a reference DNA sample known to have two copies of the chromosome. If an extra copy is present in the test sample, the signals are expected to be 1.5 times the intensities of the respective probes from the reference. If only one copy is present the proportion is expected to be 0.5. If the sample has two copies, the relative probe strengths are expected to be equal.
Dosage quotient analysis
Dosage quotient analysis is the usual method of interpreting MLPA data. If a and b are the signals from two amplicons in the patient sample, and A and B are the corresponding amplicons in the experimental control, then the dosage quotient DQ = (a/b) / (A/B). Although dosage quotients may be calculated for any pair of amplicons, it is usually the case that one of the pair is an internal reference probe.Advantages of MLPA
MLPA facilitates the amplification and detection of multiple targets with a single primer pair. In a standard multiplex PCR reaction, each fragment needs a unique amplifying primer pair. These primers being present in a large quantity result in various problems such as dimerization and false priming. With MLPA, amplification of probes can be achieved. Thus, many sequences (up to 40) can be amplified and quantified using just a single primer pair. MLPA reaction is fast, cheap and very simple to perform.Applications of MLPA
MLPA has a variety of applications including detection of mutationMutation
In molecular biology and genetics, mutations are changes in a genomic sequence: the DNA sequence of a cell's genome or the DNA or RNA sequence of a virus. They can be defined as sudden and spontaneous changes in the cell. Mutations are caused by radiation, viruses, transposons and mutagenic...
s and single nucleotide polymorphism
Single nucleotide polymorphism
A single-nucleotide polymorphism is a DNA sequence variation occurring when a single nucleotide — A, T, C or G — in the genome differs between members of a biological species or paired chromosomes in an individual...
s, analysis of DNA methylation
Methylation
In the chemical sciences, methylation denotes the addition of a methyl group to a substrate or the substitution of an atom or group by a methyl group. Methylation is a form of alkylation with, to be specific, a methyl group, rather than a larger carbon chain, replacing a hydrogen atom...
, relative mRNA quantification, chromosomal characterisation of cell lines and tissue samples, detection of gene copy number,detection of duplications and deletions in human cancer
Cancer
Cancer , known medically as a malignant neoplasm, is a large group of different diseases, all involving unregulated cell growth. In cancer, cells divide and grow uncontrollably, forming malignant tumors, and invade nearby parts of the body. The cancer may also spread to more distant parts of the...
predisposition genes such as BRCA1, BRCA2, hMLH1 and hMSH2 and aneuploidy
Aneuploidy
Aneuploidy is an abnormal number of chromosomes, and is a type of chromosome abnormality. An extra or missing chromosome is a common cause of genetic disorders . Some cancer cells also have abnormal numbers of chromosomes. Aneuploidy occurs during cell division when the chromosomes do not separate...
determination. MLPA has potential application in prenatal diagnosis
Prenatal diagnosis
Prenatal diagnosis or prenatal screening is testing for diseases or conditions in a fetus or embryo before it is born. The aim is to detect birth defects such as neural tube defects, Down syndrome, chromosome abnormalities, genetic diseases and other conditions, such as spina bifida, cleft palate,...
both invasive and noninvasive.
External links
- MLPA Products & Resources
- Applied Biosystems GeneMapper Software v4.0 fragment analysis software
- SoftGenetics GeneMarker fragment analysis software
- Conventional MLPA analysis
- Regression-enhanced MLPA analysis
- Dosage analysis examples
- Further applications of MLPA
- Special non-invasive advances in fetal and neonatal evaluation network (SAFE)
- SAFE Network non-invasive prenatal diagnosis FAQs page

