
Nucleic acid thermodynamics
Encyclopedia
Nucleic acid thermodynamics is the study of the thermodynamics
of nucleic acid
molecules, or how temperature
affects nucleic acid structure
. For multiple copies of DNA molecules, the melting temperature (Tm) is defined as the temperature at which half of the DNA strands are in the double-helical
state and half are in the random coil
states. The melting temperature depends on both the length of the molecule, and the specific nucleotide
sequence composition of that molecule.
s into a single hybrid, which in the case of two strands is referred to as a duplex
. Oligonucleotide
s, DNA
, or RNA
will bind to their complement under normal conditions, so two perfectly complementary strands will bind to each other readily. In order to reduce the diversity and obtain the most energetically preferred hybrids, a technique called annealing is used in laboratory practice. However, due to the different molecular geometries of the nucleotides, a single inconsistency between the two strands will make binding between them less energetically favorable. Measuring the effects of base incompatibility by quantifying the rate at which two strands anneal can provide information as to the similarity in base sequence between the two strands being annealed. The hybrids may be dissociated by thermal denaturation
, also referred to as melting. Here, the solution of hybrids is heated to break the hydrogen bond
s between nucleic bases, after which the two strands separate. In the absence of external negative factors, the processes of hybridization and melting may be repeated in succession indefinitely, which lays the ground for polymerase chain reaction
.
Most commonly, the pairs of nucleic bases A=T and G≡C are formed, of which the latter is more stable.
.
The process of DNA denaturation can be used to analyze some aspects of DNA. Because cytosine / guanine base-pairing is generally stronger than adenosine / thymine base-pairing, the amount of cytosine and guanine in a genome (called the "GC content") can be estimated by measuring the temperature at which the genomic DNA melts. Higher temperatures are associated with high GC content.
DNA denaturation can also be used to detect sequence differences between two different DNA sequences. DNA is heated and denatured into single-stranded state, and the mixture is cooled to allow strands to rehybridize. Hybrid molecules are formed between similar sequences and any differences between those sequences will result in a disruption of the base-pairing. On a genomic scale, the method has been used by researchers to estimate the genetic distance
between two species, a process known as DNA-DNA hybridization. In the context of a single isolated region of DNA, denaturing gradient gels and temperature gradient gels can be used to detect the presence of small mismatches between two sequences, a process known as temperature gradient gel electrophoresis
.
Methods of DNA analysis based on melting temperature have the disadvantage of being proxies for studying the underlying sequence; DNA sequencing
is generally considered a more accurate method.
The process of DNA melting is also used in molecular biology techniques, notably in the polymerase chain reaction
(PCR). Although the temperature of DNA melting is not diagnostic in the technique, methods for estimating Tm are important for determining the appropriate temperatures to use in a protocol. DNA melting temperatures can also be used as a proxy for equalizing the hybridization strengths of a set of molecules, e.g. the oligonucleotide probes of DNA microarray
s.
, means for DNA
or RNA
to pair by hydrogen bond
s to a complementary sequence
, forming a double-stranded polynucleotide
. The term is often used to describe the binding of a DNA probe, or the binding of a primer
to a DNA strand during a polymerase chain reaction
(PCR). The term is also often used to describe the reformation (renaturation) of complementary strands that were separated by heat (thermally denatured).
Proteins such as RAD52
can help DNA anneal.
Some formulas are more accurate in predicting melting temperatures of DNA duplexes.
One problem in nucleic acid thermodynamics is to determine the thermodynamic parameters for forming double-stranded nucleic acid AB from single-stranded nucleic acids A and B.
The equilibrium constant for this reaction is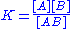 . According to thermodynamics
. According to thermodynamics
, the relation between free energy, ΔG, and K is ΔG° = -RTln K, where R is the ideal gas law constant, and T is the kelvin temperature of the reaction. This gives, for the nucleic acid system,
 .
.
The melting temperature, Tm, occurs when half of the double-stranded nucleic acid has dissociated. If no additional nucleic acids are present, then [A], [B], and [AB] will be equal, and equal to half the initial concentration of double-stranded nucleic acid, [AB]initial. This gives an expression for the melting point of a nucleic acid duplex of
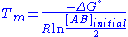 .
.
Because ΔG° = ΔH° -TΔS°, Tm is also given by
 .
.
The terms ΔH° and ΔS° are usually given for the association and not the dissociation reaction (see the nearest-neighbor method for example). This formula then turns into :
 , where [B]total < [A]total.
, where [B]total < [A]total.
This equation is based on the assumption that only two states are involved in melting: the double stranded state and the random-coil state. However, nucleic acids may melt several intermediate states. To account for such complicated behavior, the methods of statistical mechanics
must be used.
Some of these parameters can be determined using the nearest-neighbor method. The interaction between bases on different strands depends somewhat on the neighboring bases. Instead of treating a DNA helix as a string of interactions between base pairs, the nearest-neighbor model treats a DNA helix as a string of interactions between 'neighboring' base pairs. So, for example, the DNA shown below has nearest-neighbor interactions indicated by the arrows.
The free energy of forming this DNA from the individual strands, ΔG°, is represented (at 37°C) as
ΔG°37(predicted) = ΔG°37(CG initiation) + ΔG°37(CG/GC) + ΔG°37(GT/CA) + ΔG°37(TT/AA) + ΔG°37(TG/AC) + ΔG°37(GA/CT) + ΔG°37(AT initiation)
The first term represents the free energy of the first base pair, CG, in the absence of a nearest neighbor. The second term includes both the free energy of formation of the second base pair, GC, and stacking interaction between this base pair and the previous base pair. The remaining terms are similarly defined. In general, the free energy of forming a nucleic acid duplex is
 .
.
Each ΔG° term has enthalpic, ΔH°, and entropic, ΔS°, parameters, so the change in free energy is also given by
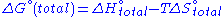 .
.
Values of ΔH° and ΔS° have been determined for the ten possible pairs of interactions. These are given in Table 1, along with the value of ΔG° calculated at 37°C. Using these values, the value of ΔG37° for the DNA helix shown above is calculated to be -22.4 kJ/mol. The experimental value is -21.8 kJ/mol.
The parameters associated with the ten groups of neighbors shown in table 1 are determined from melting points of short oligonucleotide duplexes. Curiously, it works out that only eight of the ten groups are independent. A more realistic way of modeling the behavior of nucleic acids would seem to be to have parameters that depend on the neighboring groups on both sides of a nucleotide, giving a table with entries like "TCG/AGC". However, this would involve around 32 groups; the number of experiments needed to get reliable data for so many groups would be considerable. Because the predictions from the nearest neighbor method agree reasonably well with experimental results, the extra effort required to develop a different model may not be justifiable.
Thermodynamics
Thermodynamics is a physical science that studies the effects on material bodies, and on radiation in regions of space, of transfer of heat and of work done on or by the bodies or radiation...
of nucleic acid
Nucleic acid
Nucleic acids are biological molecules essential for life, and include DNA and RNA . Together with proteins, nucleic acids make up the most important macromolecules; each is found in abundance in all living things, where they function in encoding, transmitting and expressing genetic information...
molecules, or how temperature
Temperature
Temperature is a physical property of matter that quantitatively expresses the common notions of hot and cold. Objects of low temperature are cold, while various degrees of higher temperatures are referred to as warm or hot...
affects nucleic acid structure
Nucleic acid structure
Nucleic acid structure refers to the structure of nucleic acids such as DNA and RNA It is often divided into four different levels:* Primary structure—the raw sequence of nucleobases of each of the component DNA strands;...
. For multiple copies of DNA molecules, the melting temperature (Tm) is defined as the temperature at which half of the DNA strands are in the double-helical
Nucleic acid double helix
In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA and RNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, and is a fundamental component in determining its...
state and half are in the random coil
Random coil
A random coil is a polymer conformation where the monomer subunits are oriented randomly while still being bonded to adjacent units. It is not one specific shape, but a statistical distribution of shapes for all the chains in a population of macromolecules...
states. The melting temperature depends on both the length of the molecule, and the specific nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
sequence composition of that molecule.
Hybridization
Hybridization is the process of establishing a non-covalent, sequence-specific interaction between two or more complementary strands of nucleic acidNucleic acid
Nucleic acids are biological molecules essential for life, and include DNA and RNA . Together with proteins, nucleic acids make up the most important macromolecules; each is found in abundance in all living things, where they function in encoding, transmitting and expressing genetic information...
s into a single hybrid, which in the case of two strands is referred to as a duplex
Nucleic acid double helix
In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA and RNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, and is a fundamental component in determining its...
. Oligonucleotide
Oligonucleotide
An oligonucleotide is a short nucleic acid polymer, typically with fifty or fewer bases. Although they can be formed by bond cleavage of longer segments, they are now more commonly synthesized, in a sequence-specific manner, from individual nucleoside phosphoramidites...
s, DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
, or RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
will bind to their complement under normal conditions, so two perfectly complementary strands will bind to each other readily. In order to reduce the diversity and obtain the most energetically preferred hybrids, a technique called annealing is used in laboratory practice. However, due to the different molecular geometries of the nucleotides, a single inconsistency between the two strands will make binding between them less energetically favorable. Measuring the effects of base incompatibility by quantifying the rate at which two strands anneal can provide information as to the similarity in base sequence between the two strands being annealed. The hybrids may be dissociated by thermal denaturation
Denaturation (biochemistry)
Denaturation is a process in which proteins or nucleic acids lose their tertiary structure and secondary structure by application of some external stress or compound, such as a strong acid or base, a concentrated inorganic salt, an organic solvent , or heat...
, also referred to as melting. Here, the solution of hybrids is heated to break the hydrogen bond
Hydrogen bond
A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond...
s between nucleic bases, after which the two strands separate. In the absence of external negative factors, the processes of hybridization and melting may be repeated in succession indefinitely, which lays the ground for polymerase chain reaction
Polymerase chain reaction
The polymerase chain reaction is a scientific technique in molecular biology to amplify a single or a few copies of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence....
.
Most commonly, the pairs of nucleic bases A=T and G≡C are formed, of which the latter is more stable.
Denaturation
DNA denaturation, also called DNA melting, is the process by which double-stranded deoxyribonucleic acid unwinds and separates into single-stranded strands through the breaking of hydrogen bonding between the bases. Both terms are used to refer to the process as it occurs when a mixture is heated, although "denaturation" can also refer to the separation of DNA strands induced by chemicals like ureaUrea
Urea or carbamide is an organic compound with the chemical formula CO2. The molecule has two —NH2 groups joined by a carbonyl functional group....
.
The process of DNA denaturation can be used to analyze some aspects of DNA. Because cytosine / guanine base-pairing is generally stronger than adenosine / thymine base-pairing, the amount of cytosine and guanine in a genome (called the "GC content") can be estimated by measuring the temperature at which the genomic DNA melts. Higher temperatures are associated with high GC content.
DNA denaturation can also be used to detect sequence differences between two different DNA sequences. DNA is heated and denatured into single-stranded state, and the mixture is cooled to allow strands to rehybridize. Hybrid molecules are formed between similar sequences and any differences between those sequences will result in a disruption of the base-pairing. On a genomic scale, the method has been used by researchers to estimate the genetic distance
Genetic distance
Genetic distance refers to the genetic divergence between species or between populations within a species. It is measured by a variety of parameters. Smaller genetic distances indicate a close genetic relationship whereas large genetic distances indicate a more distant genetic relationship...
between two species, a process known as DNA-DNA hybridization. In the context of a single isolated region of DNA, denaturing gradient gels and temperature gradient gels can be used to detect the presence of small mismatches between two sequences, a process known as temperature gradient gel electrophoresis
Temperature gradient gel electrophoresis
Temperature Gradient Gel Electrophoresis and Denaturing Gradient Gel Electrophoresis are forms of electrophoresis which use either a temperature or chemical gradient to denature the sample as it moves across an acrylamide gel. TGGE and DGGE can be applied to nucleic acids such as DNA and RNA,...
.
Methods of DNA analysis based on melting temperature have the disadvantage of being proxies for studying the underlying sequence; DNA sequencing
DNA sequencing
DNA sequencing includes several methods and technologies that are used for determining the order of the nucleotide bases—adenine, guanine, cytosine, and thymine—in a molecule of DNA....
is generally considered a more accurate method.
The process of DNA melting is also used in molecular biology techniques, notably in the polymerase chain reaction
Polymerase chain reaction
The polymerase chain reaction is a scientific technique in molecular biology to amplify a single or a few copies of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence....
(PCR). Although the temperature of DNA melting is not diagnostic in the technique, methods for estimating Tm are important for determining the appropriate temperatures to use in a protocol. DNA melting temperatures can also be used as a proxy for equalizing the hybridization strengths of a set of molecules, e.g. the oligonucleotide probes of DNA microarray
DNA microarray
A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome...
s.
Annealing
Annealing, in geneticsGenetics
Genetics , a discipline of biology, is the science of genes, heredity, and variation in living organisms....
, means for DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
or RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
to pair by hydrogen bond
Hydrogen bond
A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond...
s to a complementary sequence
Complementarity (molecular biology)
In molecular biology, complementarity is a property of double-stranded nucleic acids such as DNA, as well as DNA:RNA duplexes. Each strand is complementary to the other in that the base pairs between them are non-covalently connected via two or three hydrogen bonds...
, forming a double-stranded polynucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
. The term is often used to describe the binding of a DNA probe, or the binding of a primer
Primer (molecular biology)
A primer is a strand of nucleic acid that serves as a starting point for DNA synthesis. They are required for DNA replication because the enzymes that catalyze this process, DNA polymerases, can only add new nucleotides to an existing strand of DNA...
to a DNA strand during a polymerase chain reaction
Polymerase chain reaction
The polymerase chain reaction is a scientific technique in molecular biology to amplify a single or a few copies of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence....
(PCR). The term is also often used to describe the reformation (renaturation) of complementary strands that were separated by heat (thermally denatured).
Proteins such as RAD52
RAD52
RAD52 homolog , also known as RAD52, is a protein which in humans is encoded by the RAD52 gene.- Function :The protein encoded by this gene shares similarity with Saccharomyces cerevisiae Rad52, a protein important for DNA double-strand break repair and homologous recombination...
can help DNA anneal.
Methods for estimating melting temperatures
Several formulas are used to calculate Tm values.Some formulas are more accurate in predicting melting temperatures of DNA duplexes.
One problem in nucleic acid thermodynamics is to determine the thermodynamic parameters for forming double-stranded nucleic acid AB from single-stranded nucleic acids A and B.
- AB ↔ A + B
The equilibrium constant for this reaction is
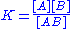 . According to thermodynamics
. According to thermodynamicsChemical equilibrium
In a chemical reaction, chemical equilibrium is the state in which the concentrations of the reactants and products have not yet changed with time. It occurs only in reversible reactions, and not in irreversible reactions. Usually, this state results when the forward reaction proceeds at the same...
, the relation between free energy, ΔG, and K is ΔG° = -RTln K, where R is the ideal gas law constant, and T is the kelvin temperature of the reaction. This gives, for the nucleic acid system,
 .
.The melting temperature, Tm, occurs when half of the double-stranded nucleic acid has dissociated. If no additional nucleic acids are present, then [A], [B], and [AB] will be equal, and equal to half the initial concentration of double-stranded nucleic acid, [AB]initial. This gives an expression for the melting point of a nucleic acid duplex of
 .
.Because ΔG° = ΔH° -TΔS°, Tm is also given by
 .
.The terms ΔH° and ΔS° are usually given for the association and not the dissociation reaction (see the nearest-neighbor method for example). This formula then turns into :
 , where [B]total < [A]total.
, where [B]total < [A]total.This equation is based on the assumption that only two states are involved in melting: the double stranded state and the random-coil state. However, nucleic acids may melt several intermediate states. To account for such complicated behavior, the methods of statistical mechanics
Statistical mechanics
Statistical mechanics or statistical thermodynamicsThe terms statistical mechanics and statistical thermodynamics are used interchangeably...
must be used.
Nearest-neighbor method
Some of these parameters can be determined using the nearest-neighbor method. The interaction between bases on different strands depends somewhat on the neighboring bases. Instead of treating a DNA helix as a string of interactions between base pairs, the nearest-neighbor model treats a DNA helix as a string of interactions between 'neighboring' base pairs. So, for example, the DNA shown below has nearest-neighbor interactions indicated by the arrows.
- ↓ ↓ ↓ ↓ ↓
- 5' C-G-T-T-G-A 3'
- 3' G-C-A-A-C-T 5'
The free energy of forming this DNA from the individual strands, ΔG°, is represented (at 37°C) as
ΔG°37(predicted) = ΔG°37(CG initiation) + ΔG°37(CG/GC) + ΔG°37(GT/CA) + ΔG°37(TT/AA) + ΔG°37(TG/AC) + ΔG°37(GA/CT) + ΔG°37(AT initiation)
The first term represents the free energy of the first base pair, CG, in the absence of a nearest neighbor. The second term includes both the free energy of formation of the second base pair, GC, and stacking interaction between this base pair and the previous base pair. The remaining terms are similarly defined. In general, the free energy of forming a nucleic acid duplex is
 .
.Each ΔG° term has enthalpic, ΔH°, and entropic, ΔS°, parameters, so the change in free energy is also given by
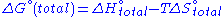 .
.Values of ΔH° and ΔS° have been determined for the ten possible pairs of interactions. These are given in Table 1, along with the value of ΔG° calculated at 37°C. Using these values, the value of ΔG37° for the DNA helix shown above is calculated to be -22.4 kJ/mol. The experimental value is -21.8 kJ/mol.
| Nearest-neighbor sequence (5'-3'/3'-5') |
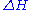 ° ° kJ/mol |
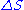 ° ° J/(mol·K) |
 °37 °37 kJ/mol |
|---|---|---|---|
| AA/TT | -33.1 | -92.9 | -4.26 |
| AT/TA | -30.1 | -85.4 | -3.67 |
| TA/AT | -30.1 | -89.1 | -2.50 |
| CA/GT | -35.6 | -95.0 | -6.12 |
| GT/CA | -35.1 | -93.7 | -6.09 |
| CT/GA | -32.6 | -87.9 | -5.40 |
| GA/CT | -34.3 | -92.9 | -5.51 |
| CG/GC | -44.4 | -113.8 | -9.07 |
| GC/CG | -41.0 | -102.1 | -9.36 |
| GG/CC | -33.5 | -83.3 | -7.66 |
| Terminal A-T base pair | 9.6 | 17.2 | 4.31 |
| Terminal G-C base pair | 0.4 | -11.7 | 4.05 |
The parameters associated with the ten groups of neighbors shown in table 1 are determined from melting points of short oligonucleotide duplexes. Curiously, it works out that only eight of the ten groups are independent. A more realistic way of modeling the behavior of nucleic acids would seem to be to have parameters that depend on the neighboring groups on both sides of a nucleotide, giving a table with entries like "TCG/AGC". However, this would involve around 32 groups; the number of experiments needed to get reliable data for so many groups would be considerable. Because the predictions from the nearest neighbor method agree reasonably well with experimental results, the extra effort required to develop a different model may not be justifiable.
See also
- DNADNADeoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
- Denaturation (biochemistry)Denaturation (biochemistry)Denaturation is a process in which proteins or nucleic acids lose their tertiary structure and secondary structure by application of some external stress or compound, such as a strong acid or base, a concentrated inorganic salt, an organic solvent , or heat...
- Melting pointMelting pointThe melting point of a solid is the temperature at which it changes state from solid to liquid. At the melting point the solid and liquid phase exist in equilibrium. The melting point of a substance depends on pressure and is usually specified at standard atmospheric pressure...
- PrimerPrimer (molecular biology)A primer is a strand of nucleic acid that serves as a starting point for DNA synthesis. They are required for DNA replication because the enzymes that catalyze this process, DNA polymerases, can only add new nucleotides to an existing strand of DNA...
for calculations of Tm - Base pairBase pairIn molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
- Polymerase chain reactionPolymerase chain reactionThe polymerase chain reaction is a scientific technique in molecular biology to amplify a single or a few copies of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence....
- Complementary DNAComplementary DNAIn genetics, complementary DNA is DNA synthesized from a messenger RNA template in a reaction catalyzed by the enzyme reverse transcriptase and the enzyme DNA polymerase. cDNA is often used to clone eukaryotic genes in prokaryotes...
- Western blotWestern blotThe western blot is a widely used analytical technique used to detect specific proteins in the given sample of tissue homogenate or extract. It uses gel electrophoresis to separate native proteins by 3-D structure or denatured proteins by the length of the polypeptide...
External links
- Tm calculations in OligoAnalyzer - Integrated DNA TechnologiesIntegrated DNA TechnologiesIntegrated DNA Technologies, Inc. , headquartered in Coralville, Iowa, is the largest supplier of custom nucleic acids in the U.S., serving the areas of academic research, biotechnology, clinical diagnostics, and pharmaceutical development...
- DNA thermodynamics calculations - Tm, melting profile, mismatches, free energy calculations
- Tm calculation - by bioPHP.org.
- http://www.promega.com/biomath/calc11.htm#disc
- Invitrogen Tm calculation
- AnnHyb Open Source software for Tm calculation using the Nearest-neighbour method
- Sigma-aldrich technical notes
- Primer3 calculation
- "Discovery of the Hybrid Helix and the First DNA-RNA Hybridization" by Alexander RichAlexander RichAlexander Rich, MD is a biologist and biophysicist. He is the William Thompson Sedgwick Professor of Biophysics at MIT and Harvard Medical School. Dr. Rich earned both an A.B. and an M.D. from Harvard University. He was a post-doc of Linus Pauling along with James Watson...
- uMelt: Melting Curve Prediction
- Nearest Neighbor Database: Provides a description of RNA-RNA interaction nearest neighbor parameters and examples of their use.

