
Protein Structure Initiative
Encyclopedia

Structural genomics
Structural genomics seeks to describe the 3-dimensional structure of every protein encoded by a given genome. This genome-based approach allows for a high-throughput method of structure determination by a combination of experimental and modeling approaches...
and contribute to understanding biological function. Funded by the U.S. National Institute of General Medical Sciences
National Institute of General Medical Sciences
The National Institute of General Medical Sciences is a part of the National Institutes of Health that primarily supports research that lays the foundation for advances in disease diagnosis, treatment and prevention...
(NIGMS), its aim is to reduce the cost and time required to determine three-dimensional protein structures and to develop techniques for solving challenging problems in structural biology, including membrane proteins. Over a dozen research centers have been supported by the PSI for work in building and maintaining high-throughput structural genomics pipelines, developing computational protein structure prediction
Protein structure prediction
Protein structure prediction is the prediction of the three-dimensional structure of a protein from its amino acid sequence — that is, the prediction of its secondary, tertiary, and quaternary structure from its primary structure. Structure prediction is fundamentally different from the inverse...
methods, organizing and disseminating information generated by the PSI, and applying high-throughput structure determination to study a broad range of important biological and biomedical problems.
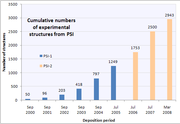
The project has been organized into three separate phases. The first phase of the Protein Structure Initiative (PSI-1) spanned from 2000 to 2005, and was dedicated to demonstrating the feasibility of high-throughput structure determination, solving unique protein structures, and preparing for a subsequent production phase. The second phase, PSI-2, focused on implementing the high-throughput structure determination methods developed in PSI-1, as well as homology modeling
Homology modeling
Homology modeling, also known as comparative modeling of protein refers to constructing an atomic-resolution model of the "target" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein...
and addressing bottlenecks like modeling membrane protein
Membrane protein
A membrane protein is a protein molecule that is attached to, or associated with the membrane of a cell or an organelle. More than half of all proteins interact with membranes.-Function:...
s. The third phase, PSI:Biology, began in 2010 and consists of networks of investigators applying high-throughput structure determination to study a broad range of biological and biomedical problems.
Phase 1
The first phase of the Protein Structure Initiative (PSI-1) lasted from June 2000 until September 2005, and had a budget of $270 million funded primarily by NIGMS with support from the National Institute of Allergy and Infectious DiseasesNational Institute of Allergy and Infectious Diseases
The National Institute of Allergy and Infectious Diseases is one of the 27 institutes and centers that make up the National Institutes of Health , an agency of the United States Department of Health and Human Services...
. PSI-1 saw the establishment of nine pilot centers focusing on structural genomics studies of a range of organisms, including Arabidopsis thaliana
Arabidopsis thaliana
Arabidopsis thaliana is a small flowering plant native to Europe, Asia, and northwestern Africa. A spring annual with a relatively short life cycle, arabidopsis is popular as a model organism in plant biology and genetics...
, Caenorhabditis elegans
Caenorhabditis elegans
Caenorhabditis elegans is a free-living, transparent nematode , about 1 mm in length, which lives in temperate soil environments. Research into the molecular and developmental biology of C. elegans was begun in 1974 by Sydney Brenner and it has since been used extensively as a model...
and Mycobacterium tuberculosis
Mycobacterium tuberculosis
Mycobacterium tuberculosis is a pathogenic bacterial species in the genus Mycobacterium and the causative agent of most cases of tuberculosis . First discovered in 1882 by Robert Koch, M...
. During this five-year period over 1,100 protein structures were determined, over 700 of which were classified as "unique" due to their < 30% sequence similarity
Sequence alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are...
with other known protein structures.
The primary goal of PSI-1, to develop methods to streamline the structure determination process, resulted in an array of technical advances. Several methods developed during PSI-1 enhanced expression
Gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA , transfer RNA or small nuclear RNA genes, the product is a functional RNA...
of recombinant
Recombinant DNA
Recombinant DNA molecules are DNA sequences that result from the use of laboratory methods to bring together genetic material from multiple sources, creating sequences that would not otherwise be found in biological organisms...
proteins in systems like Escherichia coli
Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
, Pichia pastoris
Pichia pastoris
Pichia pastoris is a species of methylotrophic yeast. Pichia is widely used for protein expression using recombinant DNA techniques. Hence it is used in biochemical and genetic research in academia and the biotechnical industry....
and insect cell lines. New streamlined approaches to cell cloning, expression and protein purification
Protein purification
Protein purification is a series of processes intended to isolate a single type of protein from a complex mixture. Protein purification is vital for the characterization of the function, structure and interactions of the protein of interest. The starting material is usually a biological tissue or...
were also introduced, in which robotics and software platforms were integrated into the protein production pipeline to minimize required manpower, increase speed, and lower costs.
Phase 2
The second phase of the Protein Structure Initiative (PSI-2) lasted from July 2005 to June 2010. Its goal was to use methods introduced in PSI-1 to determine a large number of proteins and continue development in streamlining the structural genomics pipeline. PSI-2 had a five-year budget of $325 million provided by NIGMS with support from the National Center for Research ResourcesNational Center for Research Resources
The National Center for Research Resources or NCRR, is a United States government agency. NCRR provides funding to laboratory scientists and researchers for facilities and tools in the goal of curing and treating diseases.-Organization and history:...
. By the end of this phase, the Protein Structure Initiative had solved over 4,800 protein structures; over 4,100 of these were unique.
Homology modeling
Homology modeling, also known as comparative modeling of protein refers to constructing an atomic-resolution model of the "target" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein...
centers, the Joint Center for Molecular Modeling (JCMM) and New Methods for High-Resolution Comparative Modeling (NMHRCM) were also added, as well as two resource centers, the PSI Materials Repository (PSI-MR) and the PSI Structural Biology Knowledgebase (SBKB). The TB Structural Genomics Consortium was removed from the roster of supported research centers in the transition from PSI-1 to PSI-2.
Originally launched in February 2008, the SBKB is a free resource that provides information on protein sequence and keyword searching, as well as modules describing target selection, experimental protocols, structure models, functional annotation, metrics on overall progress, and updates on structure determination technology. Like the PDB
Protein Data Bank
The Protein Data Bank is a repository for the 3-D structural data of large biological molecules, such as proteins and nucleic acids....
, it is directed by Dr. Helen M. Berman and hosted at Rutgers University
Rutgers University
Rutgers, The State University of New Jersey , is the largest institution for higher education in New Jersey, United States. It was originally chartered as Queen's College in 1766. It is the eighth-oldest college in the United States and one of the nine Colonial colleges founded before the American...
.
The PSI Materials Repository, established in 2006 at the Harvard Institute of Proteomics, stores and ships PSI-generated plasmid clones
Plasmid
In microbiology and genetics, a plasmid is a DNA molecule that is separate from, and can replicate independently of, the chromosomal DNA. They are double-stranded and, in many cases, circular...
. Clones are sequence-verified, annotated and stored in the DNASU Plasmid Repository, currently located at the Biodesign Institute at Arizona State University. As of September 2011, there are over 50,000 PSI-generated plasmid clones and empty vectors available for request through DNASU in addition to over 147,000 clones generated from non-PSI sources. Plasmids are distributed to researchers worldwide. Now called the PSI:Biology Materials Repository, this resource has a five-year budget of $5.4 million and is under the direction of Dr. Joshua LaBaer, who moved to Arizona State University in the middle of 2009, taking the PSI:Biology-MR with him.
Phase 3
The third phase of the PSI is called PSI:Biology and is intended to reflect the emphasis on the biological relevance of the work. During this phase, highly organized networks of investigators will apply the new paradigm of high-throughput structure determination, which was successfully developed during the earlier phases of the PSI, to study a broad range of important biological and biomedical problems. The network includes centers for high-throughput structure determination, centers for membrane protein structure determination, consortia for high-throughput-enabled structural biology partnerships, the SBKB and the PSI-MR. The PSI also supports two additional components: technology development for high-throughput structural biology research and technology development for protein modeling. There is an ongoing announcement for applications to establish partnerships between researchers interested in a biological problem of significant scope and researchers within the PSI:Biology network.Impact
Structural genomics
Structural genomics seeks to describe the 3-dimensional structure of every protein encoded by a given genome. This genome-based approach allows for a high-throughput method of structure determination by a combination of experimental and modeling approaches...
(SG) output was made by PSI centers. Of these PSI contributions over 20% represented new Pfam
Pfam
Pfam is a database of protein families that includes their annotations and multiple sequence alignments generated using hidden Markov models.- Features :For each family in Pfam one can:* Look at multiple alignments* View protein domain architectures...
families, compared to the non-SG average of 5%. Pfam families represent structurally distinct groups of proteins as predicted from sequenced
DNA sequencing
DNA sequencing includes several methods and technologies that are used for determining the order of the nucleotide bases—adenine, guanine, cytosine, and thymine—in a molecule of DNA....
genomes. Not targeting homologs of known structure was accomplished by using sequence comparison tools like BLAST
BLAST
In bioinformatics, Basic Local Alignment Search Tool, or BLAST, is an algorithm for comparing primary biological sequence information, such as the amino-acid sequences of different proteins or the nucleotides of DNA sequences...
and PSI-BLAST. Like the difference in novelty as determined by discovery of new Pfam families, the PSI also discovered more SCOP
Structural Classification of Proteins
The Structural Classification of Proteins database is a largely manual classification of protein structural domains based on similarities of their structures and amino acid sequences. A motivation for this classification is to determine the evolutionary relationship between proteins...
folds and superfamilies than non-SG efforts. In 2006, 16% of structures solved by the PSI represented new SCOP folds and superfamilies, while the non-SG average was 4%. Solving such novel structures reflects increased coverage of protein fold space, one of the PSI's main goals. Determining the structure a novel protein allows homology modeling
Homology modeling
Homology modeling, also known as comparative modeling of protein refers to constructing an atomic-resolution model of the "target" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein...
to more accurately predict the fold of other proteins in the same structural family.
While most of the structures solved by the four large-scale PSI centers lack functional annotation, many of the remaining PSI centers determine structures for proteins with known biological function. The TB Structural Genomics Consortium, for example, focused exclusively on functionally characterized proteins. During its term in PSI-1, it deposited structures for over 70 unique proteins from Mycobacterium tuberculosis
Mycobacterium tuberculosis
Mycobacterium tuberculosis is a pathogenic bacterial species in the genus Mycobacterium and the causative agent of most cases of tuberculosis . First discovered in 1882 by Robert Koch, M...
, which represented more than 35% of total unique M. tuberculosis structures solved through 2007. In following with its biomedical theme to increase coverage of phosphotomes, the NYSGXRC has determined structures for about 10% of all human phosphatase
Phosphatase
A phosphatase is an enzyme that removes a phosphate group from its substrate by hydrolysing phosphoric acid monoesters into a phosphate ion and a molecule with a free hydroxyl group . This action is directly opposite to that of phosphorylases and kinases, which attach phosphate groups to their...
s.
The PSI consortia have provided the overwhelming majority of targets for the Critical Assessment of Techniques for Protein Structure Prediction (CASP), a community-wide, biannual experiment to determine the state and progress of protein structure prediction
Protein structure prediction
Protein structure prediction is the prediction of the three-dimensional structure of a protein from its amino acid sequence — that is, the prediction of its secondary, tertiary, and quaternary structure from its primary structure. Structure prediction is fundamentally different from the inverse...
.
A major goal during the PSI:Biology phase is to utilize the high-throughput methods developed during the initiative's first decade to generate protein structures for functional studies, broadening the PSI's biomedical impact. It is also expected to advance knowledge and understanding of membrane proteins.
Criticism
The PSI has received notable criticism from the structural biologyStructural biology
Structural biology is a branch of molecular biology, biochemistry, and biophysics concerned with the molecular structure of biological macromolecules, especially proteins and nucleic acids, how they acquire the structures they have, and how alterations in their structures affect their function...
community. Among these charges is that the main product of the PSI – PDB
Protein Data Bank
The Protein Data Bank is a repository for the 3-D structural data of large biological molecules, such as proteins and nucleic acids....
files of proteins' atomic coordinates as determined by X-ray crystallography
X-ray crystallography
X-ray crystallography is a method of determining the arrangement of atoms within a crystal, in which a beam of X-rays strikes a crystal and causes the beam of light to spread into many specific directions. From the angles and intensities of these diffracted beams, a crystallographer can produce a...
or NMR spectroscopy
NMR spectroscopy
Nuclear magnetic resonance spectroscopy, most commonly known as NMR spectroscopy, is a research technique that exploits the magnetic properties of certain atomic nuclei to determine physical and chemical properties of atoms or the molecules in which they are contained...
– are not useful enough to biologist
Biologist
A biologist is a scientist devoted to and producing results in biology through the study of life. Typically biologists study organisms and their relationship to their environment. Biologists involved in basic research attempt to discover underlying mechanisms that govern how organisms work...
s to justify the project's $764 million cost. Critics note that money currently spent on the PSI could have otherwise funded what they consider worthier causes:
A short response to this was published:
In October 2008 the NIGMS
National Institute of General Medical Sciences
The National Institute of General Medical Sciences is a part of the National Institutes of Health that primarily supports research that lays the foundation for advances in disease diagnosis, treatment and prevention...
hosted a meeting concerning the future of structural genomics efforts and invited speakers from the PSI Advisory Committee, members of the NIGMS Advisory Council, and interested scientists who had no previous involvement with the PSI. Representatives of other genomics, proteomics, and structural genomics initiatives, as well as scientists from academia, government, and industry were also included. Based on this meeting and the subsequent recommendations from the PSI Advisory Committee, a concept-clearance document was released in January 2009 describing what a third phase of the PSI might entail. Most notable was a large emphasis on partnerships and collaborations to ensure that the majority of PSI research is focused on proteins of interest to the broader research community as well as efforts to make PSI products more accessible to the research community.
Grant applications for PSI:Biology were submitted by October 29, 2009. See Phase 3 section above.
External links
- Protein Structure Initiative
- PSI:Biology Funded Centers and Grants
- Structural Biology Knowledgebase
- PSI:Biology-Materials Repository
- Open Protein Structure Annotation Network (TOPSAN), a wikiWikiA wiki is a website that allows the creation and editing of any number of interlinked web pages via a web browser using a simplified markup language or a WYSIWYG text editor. Wikis are typically powered by wiki software and are often used collaboratively by multiple users. Examples include...
for annotation of protein structures determined by the PSI

