
Ribbon diagram
Encyclopedia
Peptide
Peptides are short polymers of amino acid monomers linked by peptide bonds. They are distinguished from proteins on the basis of size, typically containing less than 50 monomer units. The shortest peptides are dipeptides, consisting of two amino acids joined by a single peptide bond...
bonds (put together by the ribosome
Ribosome
A ribosome is a component of cells that assembles the twenty specific amino acid molecules to form the particular protein molecule determined by the nucleotide sequence of an RNA molecule....
, according to the genetic code
Genetic code
The genetic code is the set of rules by which information encoded in genetic material is translated into proteins by living cells....
as stored in DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
genes
Gênes
Gênes is the name of a département of the First French Empire in present Italy, named after the city of Genoa. It was formed in 1805, when Napoleon Bonaparte occupied the Republic of Genoa. Its capital was Genoa, and it was divided in the arrondissements of Genoa, Bobbio, Novi Ligure, Tortona and...
, transcribed into messenger RNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
, and then translated by the ribosome into the amino-acid sequence of the protein). Each amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
has one of 20 types of side chains, functional groups of different sizes and chemistries, which branch off from the backbone. Once synthesized, a protein molecule folds
Protein folding
Protein folding is the process by which a protein structure assumes its functional shape or conformation. It is the physical process by which a polypeptide folds into its characteristic and functional three-dimensional structure from random coil....
up by interactions of those sidechains: 1) making hydrogen bond
Hydrogen bond
A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond...
s between backbone peptides to form α-helix
Alpha helix
A common motif in the secondary structure of proteins, the alpha helix is a right-handed coiled or spiral conformation, in which every backbone N-H group donates a hydrogen bond to the backbone C=O group of the amino acid four residues earlier...
and β-sheet
Beta sheet
The β sheet is the second form of regular secondary structure in proteins, only somewhat less common than the alpha helix. Beta sheets consist of beta strands connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet...
secondary structures, and 2) burying the hydrophobic or non-polar sidechains on the inside away from water. The 3D shape of the folded protein produces its functional, biological properties such as enzyme
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
catalysis or specific binding of other molecules.
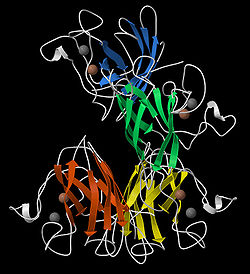
Ribbon diagrams, also known as Richardson Diagrams, are 3D
Three-dimensional space
Three-dimensional space is a geometric 3-parameters model of the physical universe in which we live. These three dimensions are commonly called length, width, and depth , although any three directions can be chosen, provided that they do not lie in the same plane.In physics and mathematics, a...
schematic representations of protein structure
Protein structure
Proteins are an important class of biological macromolecules present in all organisms. Proteins are polymers of amino acids. Classified by their physical size, proteins are nanoparticles . Each protein polymer – also known as a polypeptide – consists of a sequence formed from 20 possible L-α-amino...
and are one of the most common methods of protein depiction used today. The ribbon shows the overall path and organization of the protein backbone in 3D, and serves as a visual framework on which to hang details of the full atomic structure, such as the balls for the copper and zinc atoms at the active site of superoxide dismutase in the image at the right. Ribbon diagrams are generated by interpolating a smooth curve through the polypeptide backbone. α-helices
Alpha helix
A common motif in the secondary structure of proteins, the alpha helix is a right-handed coiled or spiral conformation, in which every backbone N-H group donates a hydrogen bond to the backbone C=O group of the amino acid four residues earlier...
are shown as coiled ribbons or thick tubes, β-strands as arrows, and lines or thin tubes for non-repetitive coils or loops. The direction of the polypeptide chain
Peptide
Peptides are short polymers of amino acid monomers linked by peptide bonds. They are distinguished from proteins on the basis of size, typically containing less than 50 monomer units. The shortest peptides are dipeptides, consisting of two amino acids joined by a single peptide bond...
is shown locally by the arrows, and may be indicated overall by a colour ramp along the length of the ribbon.
Ribbon diagrams are simple, yet powerful, in expressing the visual basics of a molecular structure (twist, fold and unfold). This method has successfully portrayed the overall organization of the protein structure, reflecting its 3-dimensional information, and allowing for better understanding of a complex object both by the expert structural biologists and also by other scientists, students, and the general public.
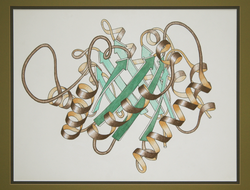
History
Originally conceived by Jane S. RichardsonJane S. Richardson
Jane Shelby Richardson is an American biochemist who developed the Richardson diagram, or ribbon diagram, method of representing proteins...
in 1980 (influenced by some earlier individual illustrations, e.g., see), her hand-drawn ribbon diagrams were the first schematics of 3D protein structure to be produced systematically, to illustrate a classification of protein structures for an article in Advances in Protein Chemistry (now available in annotated form on-line at Anatax). These drawings were made in pen on tracing paper over a printout of a Cα trace of the atomic coordinates; they preserved positions, smoothed the backbone path, and incorporated small local shifts to disambiguate the visual appearance. As well as the TIM ribbon drawing at the right, other hand-drawn examples are for prealbumin, flavodoxin, and Cu,Zn superoxide dismutase.
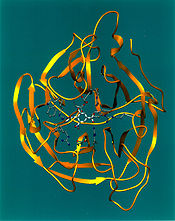
Arthur M. Lesk
Arthur M. Lesk, is an accomplished protein science researcher, who is currently a professor of biochemistry and molecular biology at the Pennsylvania State University, University Park.- Profile :...
and co-workers first enabled automatic generation of ribbon diagrams through a computational implementation that uses Protein Data Bank files as input. This conceptually simple algorithm
Algorithm
In mathematics and computer science, an algorithm is an effective method expressed as a finite list of well-defined instructions for calculating a function. Algorithms are used for calculation, data processing, and automated reasoning...
fit cubic polynomial B-spline
B-spline
In the mathematical subfield of numerical analysis, a B-spline is a spline function that has minimal support with respect to a given degree, smoothness, and domain partition. B-splines were investigated as early as the nineteenth century by Nikolai Lobachevsky...
curves to the peptide planes. Most modern graphics systems provide either B-splines or Hermite spline
Hermite spline
In the mathematical subfield of numerical analysis, a Hermite spline is a spline curve where each polynomial of the spline is in Hermite form.-See also:*Cubic Hermite spline*Hermite polynomials*Hermite interpolation...
s as a basic drawing primitive. One type of spline implementation passes through each Cα guide point, producing an exact but choppy curve. Both hand-drawn and most computer ribbons (such as those shown here) are smoothed over about 4 successive guide points (usually the peptide midpoint), to produce a more visually pleasing and understandable representation. In order to give the right radius for helical spirals while preserving smooth β-strands, the splines can be modified by offsets proportional to local curvature, as first developed by Mike Carson for his Ribbons program (figure at right) and later adapted by other molecular graphics software, such as the open-source Mage program for kinemage
Kinemage
A kinemage is an interactive graphic scientific illustration. It often is used to visualize molecules, especially proteins and nucleic acids, although it can also represent other types of 3-dimensional data...
graphics that produced the ribbon image at top right (other examples: 1xk8 trimer and DNA polymerase).
Since their inception, and continuing in the present, a ribbon diagram is the single most common representation of protein structures and a very common choice of cover image for a journal or textbook.
Current computer programs
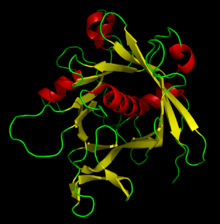
Hermite spline
In the mathematical subfield of numerical analysis, a Hermite spline is a spline curve where each polynomial of the spline is in Hermite form.-See also:*Cubic Hermite spline*Hermite polynomials*Hermite interpolation...
s to create coordinates for coils, turns, strands and helices. The curve passes through all its control points (Cα atoms) guided by direction vectors. The program was built on the basis of traditional molecular graphics by Arthur M. Lesk
Arthur M. Lesk
Arthur M. Lesk, is an accomplished protein science researcher, who is currently a professor of biochemistry and molecular biology at the Pennsylvania State University, University Park.- Profile :...
, Karl Hardman, and John Priestle. Jmol
Jmol
Jmol is an open-source Java viewer for chemical structures in 3D,that does not require 3D acceleration plugins.Jmol returns a 3D representation of a molecule that may be used as a teaching tool, or for research e.g...
is an open-source Java-based viewer for browsing molecular structures on the web; it includes a simplified "cartoon" version of ribbons. Other graphics programs such as DeepView (example: urease) and MolMol (example: SH2 domain) also produce ribbon images. KiNG is the Java-based successor to Mage (examples: α-hemolysin top view and side view).
UCSF Chimera
UCSF Chimera
UCSF Chimera is an extensible program for interactive visualization and analysis of molecular structures and related data, including density maps, supramolecular assemblies, sequence alignments, docking results, trajectories, and conformational ensembles. High-quality and can be created...
is a powerful molecular modeling program that also includes visualizations
Scientific visualization
Scientific visualization is an interdisciplinary branch of science according to Friendly "primarily concerned with the visualization of three-dimensional phenomena , where the emphasis is on realistic renderings of volumes, surfaces, illumination sources, and so forth, perhaps...
such as ribbons, notable especially for the ability to combine them with contoured shapes from cryo-electron microscopy data. PyMOL
PyMOL
PyMOL is an open-source, user-sponsored, molecular visualization system created by Warren Lyford DeLano and commercialized by DeLano Scientific LLC, which is a private software company dedicated to creating useful tools that become universally accessible to scientific and educational communities...
, by Warren DeLano, is a very popular and flexible molecular graphics program (based on Python
Python (programming language)
Python is a general-purpose, high-level programming language whose design philosophy emphasizes code readability. Python claims to "[combine] remarkable power with very clear syntax", and its standard library is large and comprehensive...
) that operates in interactive mode and also produces presentation-quality 2D images for ribbon diagrams and many other representations (see figure at right, and also mabinlin and TNF-alpha).
Features of ribbon diagrams
| Secondary Structure | ||
|---|---|---|
| α-Helices | Cylindrical spiral ribbons, with ribbon plane approximately following plane of peptides. | |
| β-Strand | Arrows with thickness, about one-quarter as thick as they are wide, shows direction and twist of the strand from amino to the carboxy end. β-sheets are seen as unified, because neighboring strands twist in unison. | |
| Loops and miscellaneous | ||
| Nonrepetitive loops | Round ropes that are fatter in the foreground and thinner towards the back, following smoothed path of Cα trace. | |
| Junctions between loops and helices | Round rope that gradually flattens out into a thin helical ribbon. | |
| Other features | ||
| Polypeptide direction, NH2 and COOH termini |
Small arrows on one or both of the termini or by letters. For β-strands, the direction of the arrow is sufficient. Today, the direction of the polypeptide chain is often indicated by a colour ramp. | |
| Disulfide bonds | Interlocked SS symbol or a zigzag, like a stylized lightning stroke | |
| Prosthetic groups or inhibitors | Stick figures, or ball&stick. | |
| Metals | Spheres (e.g., see top image). | |
| Shading and colour | Shading or colour adds dimensionality to the diagram. Generally, the features at the front are the darkest, while becoming lighter and lower in contrast towards the back. | |

