Silac
Encyclopedia
Mass spectrometry
Mass spectrometry is an analytical technique that measures the mass-to-charge ratio of charged particles.It is used for determining masses of particles, for determining the elemental composition of a sample or molecule, and for elucidating the chemical structures of molecules, such as peptides and...
that detects differences in protein abundance among samples using non-radioactive isotopic labeling
Isotopic labeling
Isotopic labeling is a technique for tracking the passage of a sample of substance through a system. The substance is 'labeled' by including unusual isotopes in its chemical composition...
. It is a popular method for quantitative proteomics
Quantitative proteomics
The aim of quantitative proteomics is to obtain quantitative information about all proteins in a sample. Rather than just providing lists of proteins identified in a certain sample, quantitative proteomics yields information about differences between samples. For example, this approach can be used...
.
Procedure
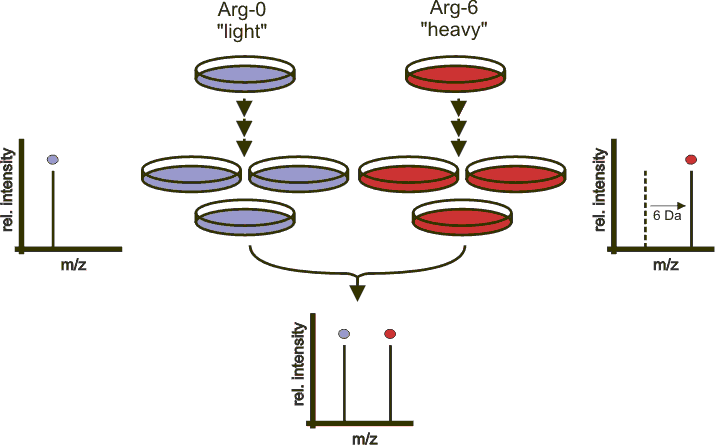
Two populations of cells are cultivated in cell cultureCell culture
Cell culture is the complex process by which cells are grown under controlled conditions. In practice, the term "cell culture" has come to refer to the culturing of cells derived from singlecellular eukaryotes, especially animal cells. However, there are also cultures of plants, fungi and microbes,...
. One of the cell populations is fed with growth medium
Growth medium
A growth medium or culture medium is a liquid or gel designed to support the growth of microorganisms or cells, or small plants like the moss Physcomitrella patens.There are different types of media for growing different types of cells....
containing normal amino acids. In contrast, the second population is fed with growth medium containing amino acids labeled with stable (non-radioactive) heavy isotope
Isotope
Isotopes are variants of atoms of a particular chemical element, which have differing numbers of neutrons. Atoms of a particular element by definition must contain the same number of protons but may have a distinct number of neutrons which differs from atom to atom, without changing the designation...
s. For example, the medium can contain arginine
Arginine
Arginine is an α-amino acid. The L-form is one of the 20 most common natural amino acids. At the level of molecular genetics, in the structure of the messenger ribonucleic acid mRNA, CGU, CGC, CGA, CGG, AGA, and AGG, are the triplets of nucleotide bases or codons that codify for arginine during...
labeled with six carbon-13
Carbon-13
Carbon-13 is a natural, stable isotope of carbon and one of the environmental isotopes. It makes up about 1.1% of all natural carbon on Earth.- Detection by mass spectrometry :...
atoms (13C) instead of the normal carbon-12
Carbon-12
Carbon-12 is the more abundant of the two stable isotopes of the element carbon, accounting for 98.89% of carbon; it contains 6 protons, 6 neutrons, and 6 electrons....
(12C). When the cells are growing in this medium, they incorporate the heavy arginine into all of their proteins. Therefore, all of the arginine containing peptides are now 6 Da heavier than their normal counterparts. The trick is that the proteins from both cell populations can be combined and analyzed together by mass spectrometry
Mass spectrometry
Mass spectrometry is an analytical technique that measures the mass-to-charge ratio of charged particles.It is used for determining masses of particles, for determining the elemental composition of a sample or molecule, and for elucidating the chemical structures of molecules, such as peptides and...
. Pairs of chemically identical peptides of different stable-isotope composition can be differentiated in a mass spectrometer owing to their mass difference. The ratio of peak intensities in the mass spectrum for such peptide pairs reflects the abundance ratio for the two proteins.
Applications
A SILAC approach involving incorporation of tyrosineTyrosine
Tyrosine or 4-hydroxyphenylalanine, is one of the 22 amino acids that are used by cells to synthesize proteins. Its codons are UAC and UAU. It is a non-essential amino acid with a polar side group...
labeled with nine carbon-13
Carbon-13
Carbon-13 is a natural, stable isotope of carbon and one of the environmental isotopes. It makes up about 1.1% of all natural carbon on Earth.- Detection by mass spectrometry :...
atoms (13C) instead of the normal carbon-12
Carbon-12
Carbon-12 is the more abundant of the two stable isotopes of the element carbon, accounting for 98.89% of carbon; it contains 6 protons, 6 neutrons, and 6 electrons....
(12C) has been utilized to study tyrosine kinase substrates in signaling pathways. SILAC has emerged as a very powerful method to study cell signaling
Cell signaling
Cell signaling is part of a complex system of communication that governs basic cellular activities and coordinates cell actions. The ability of cells to perceive and correctly respond to their microenvironment is the basis of development, tissue repair, and immunity as well as normal tissue...
, post translation modifications such as phosphorylation
Phosphorylation
Phosphorylation is the addition of a phosphate group to a protein or other organic molecule. Phosphorylation activates or deactivates many protein enzymes....
, protein–protein interaction and regulation of gene expression
Regulation of gene expression
Gene modulation redirects here. For information on therapeutic regulation of gene expression, see therapeutic gene modulation.Regulation of gene expression includes the processes that cells and viruses use to regulate the way that the information in genes is turned into gene products...
. In addition, SILAC has become an important method in secretomics
Secretomics
Secretomics is a subset of proteomics in which all of the secreted proteins of a cell, tissue, or organism are analyzed. Secreted proteins are involved in a variety of physiological processes, including cell signaling and matrix remodeling, but are also integral to invasion and metastasis of...
, the global study of secreted proteins
Secretory protein
A secretory protein is any protein, whether it be endocrine or exocrine, which is secreted by a cell. Secretory proteins include many hormones, enzymes, toxins, and antimicrobial peptides.Secretory proteins are synthesized in endoplasmic reticulum....
and secretory pathways. It can be used to distinguish between proteins secreted by cells in culture and serum contaminants. Standardized protocols of SILAC for various application have also been published.
Pulsed SILAC
Pulsed SILAC (pSILAC) is a variation of the SILAC method where the labelled amino acids are added to the growth medium for only a short period of time. This allows monitoring differences in de novo protein production rather than raw concentration.See also
- Isotope-coded affinity tag (ICAT)Isotope-coded affinity tagIsotope-coded affinity tags are a gel-free method for quantitative proteomics that relies on chemical labeling reagents. These chemical probes consist of three general elements: a reactive group capable of labeling a defined amino acid side chain , an isotopically coded linker, and a tag for the...
- Isobaric labelingIsobaric labelingIsobaric labeling is a mass spectrometry strategy used in quantitative proteomics. Peptides or proteins are labeled with various chemical groups that are isobaric, or the same in mass, but which fragment during tandem mass spectrometry to yield reporter ions of different mass...
- Tandem mass tags (TMT)Tandem mass tagsTandem mass tags are chemical labels used for mass spectrometry -based quantification and identification of biological macromolecules such as proteins, peptides and nucleic acids. TMT belongs to a family of reagents referred to as isobaric mass tags...
- Isobaric tags for relative and absolute quantitation (iTRAQ)ITRAQIsobaric tags for relative and absolute quantitation are a non-gel-based technique used to quantify proteins from different sources in a single experiment. It uses isotope-coded covalent tags...
- Tandem mass tags (TMT)
- Label-free quantificationLabel-free quantificationLabel-free quantification is a method in mass spectrometry that aims to determine the differential expression level of proteins in two or more biological samples...

