Transposase
Encyclopedia
Transposase is an enzyme
that binds to the ends of a transposon
and catalyzes the movement of the transposon
to another part of the genome by a cut and paste mechanism or a replicative transposition mechanism.
The word "transposase" was first coined by the individuals who cloned the enzyme required for tranposition of the Tn3 transposon. The existence of transposons was postulated in the late 1940s by Barbara McClintock
, who was studying the inheritance of maize, but the actual molecular basis for transposition was described by later groups. McClintock discovered that pieces of the chromosomes changed their position, jumping from one chromosome to another. The repositioning of these transposons (which coded for color) allowed other genes for pigment to be expressed. Transposition in maize causes changes in color; however, in other organisms, such as bacteria, it can cause antibiotic resistance. Transposition is also important in creating genetic diversity within species and adaptability to changing living conditions. During the course of human evolution, as much as 40% of the human genome has moved around via methods such as transposition of transposons.
Transposases are classified under EC number
EC 2.7.7.
superfamily of proteins which includes retroviral integrase
s. Tn5 can be found in Shewanella
and Escherichia
bacteria. The transposon codes for antibiotic resistance to kanamycin
and other aminoglycoside antibiotics.
Tn5 and other transposases are notably inactive. Because DNA transposition events are inherently mutagenic, the low activity of transposases is necessary to reduce the risk of causing a fatal mutation in the host, and thus eliminating the transposable element. One of the reasons Tn5 is so unreactive is because the N- and C-termini are located in relatively close proximity to one another and tend to inhibit each other. This was elucidated by the characterization of several mutations which resulted in hyperactive forms of transposases. One such mutation, L372P, is a mutation of amino acid 372 in the Tn5 transposase. This amino acid is generally a leucine residue in the middle of an alpha helix. When this leucine is replaced with a proline residue the alpha helix is broken, introducing a conformational change to the C-Terminal domain, separating it from the N-Terminal domain enough to promote higher activity of the protein. The transposition of a transposon often needs only three pieces: the transposon, the transposase enzyme, and the target DNA for the insertion of the transposon. This is the case with Tn5, which uses a cut-and-paste mechanism for moving around transposons.
Tn5 and most other transposases contain a DDE motif, which is the active site that catalyzes the movement of the transposon. Aspartate-97, Aspartate-188, and Glutamate-326 make up the active site, which is a triad of acidic residues. The DDE motif is said to coordinate divalent metal ions, most often magnesium and manganese, which are important in the catalytic reaction. Because transposase is incredibly inactive, the DDE region is mutated so that the transposase becomes hyperactive and catalyzes the movement of the transposon. The glutamate is transformed into an aspartate and the two asparates into glutamates. Through this mutation, the study of Tn5 becomes possible, but some steps in the catalytic process are lost as a result.
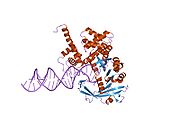 There are several steps which catalyze the movement of the transposon, including Tnp binding, synapsis (the creation of a synaptic complex), cleavage, target capture, and strand transfer. Transposase then binds to the DNA strand and creates a clamp over the transposon end of the DNA and inserts into the active site. Once the transposase binds to the transposon, it produces a synaptic complex in which two transposases are bound in a cis/trans relationship with the transposon.
There are several steps which catalyze the movement of the transposon, including Tnp binding, synapsis (the creation of a synaptic complex), cleavage, target capture, and strand transfer. Transposase then binds to the DNA strand and creates a clamp over the transposon end of the DNA and inserts into the active site. Once the transposase binds to the transposon, it produces a synaptic complex in which two transposases are bound in a cis/trans relationship with the transposon.
In cleavage, the magnesium ions activate oxygen from water molecules and expose them to nucleophilic attack. This allows the water molecules to nick the 3' strands on both ends and create a hairpin formation, which separates the transposon from the donor DNA. Next, the transposase moves the transposon to a suitable location. Not much is known about the target capture, although there is a sequence bias which has not yet been determined. After target capture, the transposase attacks the target DNA nine base pairs apart, resulting in the integration of the transposon into the target DNA.
As mentioned before, due to the mutations of the DDE, some steps of the process are lost—for example, when this experiment is performed in vitro, and SDS heat treatment denatures the transposase. However, it is still uncertain what happens to the transposase in vivo.
The study of transposase Tn5 is of general importance because of its similarities to HIV
-1 and other retroviral diseases. By studying Tn5, much can also be discovered about other transposases and their activities.
. SB transposase belongs to the DD[E/D] family of transposases, which in turn belong to a large superfamily of polynucleotidyl transferases that includes RNase H, RuvC Holliday resolvase, RAG proteins, and retroviral integrases. The SB system is used primarily in vertebrate animals for gene transfer, including gene therapy, and gene discovery The recently engineered SB100X is an especially powerful enzyme that directs the highest levels of transposon integration yet developed
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
that binds to the ends of a transposon
Transposon
Transposable elements are sequences of DNA that can move or transpose themselves to new positions within the genome of a single cell. The mechanism of transposition can be either "copy and paste" or "cut and paste". Transposition can create phenotypically significant mutations and alter the cell's...
and catalyzes the movement of the transposon
Transposon
Transposable elements are sequences of DNA that can move or transpose themselves to new positions within the genome of a single cell. The mechanism of transposition can be either "copy and paste" or "cut and paste". Transposition can create phenotypically significant mutations and alter the cell's...
to another part of the genome by a cut and paste mechanism or a replicative transposition mechanism.
The word "transposase" was first coined by the individuals who cloned the enzyme required for tranposition of the Tn3 transposon. The existence of transposons was postulated in the late 1940s by Barbara McClintock
Barbara McClintock
Barbara McClintock , the 1983 Nobel Laureate in Physiology or Medicine, was an American scientist and one of the world's most distinguished cytogeneticists. McClintock received her PhD in botany from Cornell University in 1927, where she was a leader in the development of maize cytogenetics...
, who was studying the inheritance of maize, but the actual molecular basis for transposition was described by later groups. McClintock discovered that pieces of the chromosomes changed their position, jumping from one chromosome to another. The repositioning of these transposons (which coded for color) allowed other genes for pigment to be expressed. Transposition in maize causes changes in color; however, in other organisms, such as bacteria, it can cause antibiotic resistance. Transposition is also important in creating genetic diversity within species and adaptability to changing living conditions. During the course of human evolution, as much as 40% of the human genome has moved around via methods such as transposition of transposons.
Transposases are classified under EC number
EC number
The Enzyme Commission number is a numerical classification scheme for enzymes, based on the chemical reactions they catalyze....
EC 2.7.7.
Transposase Tn5
Transposase Tn5 is a member of the RNaseRibonuclease
Ribonuclease is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 and 3.1 classes of enzymes.-Function:All organisms studied contain...
superfamily of proteins which includes retroviral integrase
Integrase
Retroviral integrase is an enzyme produced by a retrovirus that enables its genetic material to be integrated into the DNA of the infected cell...
s. Tn5 can be found in Shewanella
Shewanella
Shewanella is the sole genus included in the Shewanellaceae family of marine bacteria. Shewanella is a marine bacterium capable of modifying metals, by saturating them with electrons causing the metal to expand and soften, allowing them to process it, which in turn releases an electrical charge...
and Escherichia
Escherichia
Escherichia is a genus of Gram-negative, non-spore forming, facultatively anaerobic, rod-shaped bacteria from the family Enterobacteriaceae. In those species which are inhabitants of the gastrointestinal tracts of warm-blooded animals, Escherichia species provide a portion of the...
bacteria. The transposon codes for antibiotic resistance to kanamycin
Kanamycin
Kanamycin sulfate is an aminoglycoside antibiotic, available in oral, intravenous, and intramuscular forms, and used to treat a wide variety of infections. Kanamycin is isolated from Streptomyces kanamyceticus.-Mechanism:...
and other aminoglycoside antibiotics.
Tn5 and other transposases are notably inactive. Because DNA transposition events are inherently mutagenic, the low activity of transposases is necessary to reduce the risk of causing a fatal mutation in the host, and thus eliminating the transposable element. One of the reasons Tn5 is so unreactive is because the N- and C-termini are located in relatively close proximity to one another and tend to inhibit each other. This was elucidated by the characterization of several mutations which resulted in hyperactive forms of transposases. One such mutation, L372P, is a mutation of amino acid 372 in the Tn5 transposase. This amino acid is generally a leucine residue in the middle of an alpha helix. When this leucine is replaced with a proline residue the alpha helix is broken, introducing a conformational change to the C-Terminal domain, separating it from the N-Terminal domain enough to promote higher activity of the protein. The transposition of a transposon often needs only three pieces: the transposon, the transposase enzyme, and the target DNA for the insertion of the transposon. This is the case with Tn5, which uses a cut-and-paste mechanism for moving around transposons.
Tn5 and most other transposases contain a DDE motif, which is the active site that catalyzes the movement of the transposon. Aspartate-97, Aspartate-188, and Glutamate-326 make up the active site, which is a triad of acidic residues. The DDE motif is said to coordinate divalent metal ions, most often magnesium and manganese, which are important in the catalytic reaction. Because transposase is incredibly inactive, the DDE region is mutated so that the transposase becomes hyperactive and catalyzes the movement of the transposon. The glutamate is transformed into an aspartate and the two asparates into glutamates. Through this mutation, the study of Tn5 becomes possible, but some steps in the catalytic process are lost as a result.

In cleavage, the magnesium ions activate oxygen from water molecules and expose them to nucleophilic attack. This allows the water molecules to nick the 3' strands on both ends and create a hairpin formation, which separates the transposon from the donor DNA. Next, the transposase moves the transposon to a suitable location. Not much is known about the target capture, although there is a sequence bias which has not yet been determined. After target capture, the transposase attacks the target DNA nine base pairs apart, resulting in the integration of the transposon into the target DNA.
As mentioned before, due to the mutations of the DDE, some steps of the process are lost—for example, when this experiment is performed in vitro, and SDS heat treatment denatures the transposase. However, it is still uncertain what happens to the transposase in vivo.
The study of transposase Tn5 is of general importance because of its similarities to HIV
HIV
Human immunodeficiency virus is a lentivirus that causes acquired immunodeficiency syndrome , a condition in humans in which progressive failure of the immune system allows life-threatening opportunistic infections and cancers to thrive...
-1 and other retroviral diseases. By studying Tn5, much can also be discovered about other transposases and their activities.
Sleeping Beauty transposase
The Sleeping Beauty (SB) transpose is the recombinase that drives the Sleeping Beauty transposon systemSleeping Beauty transposon system
The Sleeping Beauty transposon system is a synthetic DNA transposon that was constructed to introduce precisely defined DNA sequences into the chromosomes of vertebrate animals for the purposes of introducing new traits and to discover new genes and their functions.-Mechanism of Action:The Sleeping...
. SB transposase belongs to the DD[E/D] family of transposases, which in turn belong to a large superfamily of polynucleotidyl transferases that includes RNase H, RuvC Holliday resolvase, RAG proteins, and retroviral integrases. The SB system is used primarily in vertebrate animals for gene transfer, including gene therapy, and gene discovery The recently engineered SB100X is an especially powerful enzyme that directs the highest levels of transposon integration yet developed

