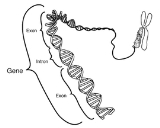
Intron
Encyclopedia
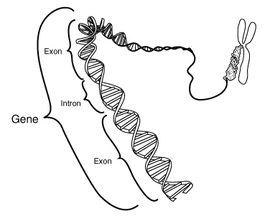
Gene
A gene is a molecular unit of heredity of a living organism. It is a name given to some stretches of DNA and RNA that code for a type of protein or for an RNA chain that has a function in the organism. Living beings depend on genes, as they specify all proteins and functional RNA chains...
that is removed by RNA splicing
RNA splicing
In molecular biology and genetics, splicing is a modification of an RNA after transcription, in which introns are removed and exons are joined. This is needed for the typical eukaryotic messenger RNA before it can be used to produce a correct protein through translation...
to generate the final mature RNA product of a gene. The term intron refers to both the DNA sequence within a gene, and the corresponding sequence in RNA transcripts. Sequences that are joined together in the final mature RNA after RNA splicing are exon
Exon
An exon is a nucleic acid sequence that is represented in the mature form of an RNA molecule either after portions of a precursor RNA have been removed by cis-splicing or when two or more precursor RNA molecules have been ligated by trans-splicing. The mature RNA molecule can be a messenger RNA...
s. Introns are found in the genes of most organisms and many viruses, and can be located in a wide range of genes, including those that generate protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s, ribosomal RNA
Ribosomal RNA
Ribosomal ribonucleic acid is the RNA component of the ribosome, the enzyme that is the site of protein synthesis in all living cells. Ribosomal RNA provides a mechanism for decoding mRNA into amino acids and interacts with tRNAs during translation by providing peptidyl transferase activity...
(rRNA), and transfer RNA
Transfer RNA
Transfer RNA is an adaptor molecule composed of RNA, typically 73 to 93 nucleotides in length, that is used in biology to bridge the three-letter genetic code in messenger RNA with the twenty-letter code of amino acids in proteins. The role of tRNA as an adaptor is best understood by...
(tRNA). When proteins are generated from intron-containing genes, RNA splicing takes place as part of the RNA processing pathway that follows transcription
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
and precedes translation.
The word intron is derived from the term intragenic region, i.e. a region inside a gene. Although introns are sometimes called intervening sequences, the term "intervening sequence" can refer to any of several families of internal nucleic acid sequences that are not present in the final gene product, including inteins, untranslated sequences (UTR), and nucleotides removed by RNA editing
RNA editing
The term RNA editing describes those molecular processes in which the information content in an RNA molecule is altered through a chemical change in the base makeup. To date, such changes have been observed in tRNA, rRNA, mRNA and microRNA molecules of eukaryotes but not prokaryotes...
, in addition to introns.
Simple illustration of an intron and RNA splicing
Introns and the process of RNA splicing can be illustrated using a text example. Consider the following quotation from Groucho MarxGroucho Marx
Julius Henry "Groucho" Marx was an American comedian and film star famed as a master of wit. His rapid-fire delivery of innuendo-laden patter earned him many admirers. He made 13 feature films with his siblings the Marx Brothers, of whom he was the third-born...
:
"Outside of a dog, a book is man's best friend. Inside of a dog it's too dark to read."
To make this text look more like a nucleic acid sequence, we'll simply delete all punctuation and make all of the text uppercase. This will be our "message".
OUTSIDEOFADOGABOOKISMANSBESTFRIENDINSIDEOFADOGITSTOODARKTOREAD
If there were a single intron within this sequence, it would look something like this:
OUTSIDEOFADOGABOOKISMANSBEGUITUITGLSAJSAKHDLAYSIOEYASHDKLSALKDN
KLASNDKLASKGDASJKBDNKNSKDKLSANDKNSKANDKNSAKNDKSAKLDHSDJGJASBDKN
SANDLNSMALVJOPDHVANVLNVKLSAKNFADNGKLHKNIUHSAFLSAFLFASFOLSANFLNK
FNKDSNGOIFSGHSKDHGKDHSFIABHFIHEHRFKHLKDHSUNSIUANIDAUBIOAYICMUSF
AVMASSTFRIENDINSIDEOFADOGITSTOODARKTOREAD
1. Identify exon and intron sequences
To remove the intron, the sequences corresponding to the intron and exons must be precisely identified. Here, the exon sequences are changed to bold. Note that misidentifying the boundaries between the exons and intron by even a single letter will result in a nonsensical message.OUTSIDEOFADOGABOOKISMANSBEGUITUITGLSAJSAKHDLAYSIOEYASHDKLSALKDN
KLASNDKLASKGDASJKBDNKNSKDKLSANDKNSKANDKNSAKNDKSAKLDHSDJGJASBDKN
SANDLNSMALVJOPDHVANVLNVKLSAKNFADNGKLHKNIUHSAFLSAFLFASFOLSANFLNK
FNKDSNGOIFSGHSKDHGKDHSFIABHFIHEHRFKHLKDHSUNSIUANIDAUBIOAYICMUSF
AVMASSTFRIENDINSIDEOFADOGITSTOODARKTOREAD
2. Cleavage at intron-exon junctions
Next, the sequence must be cut precisely at the boundaries between the exons and the intronOUTSIDEOFADOGABOOKISMANSBE
GUITUITGLSAJSAKHDLAYSIOEYASHDKLSALKDN
KLASNDKLASKGDASJKBDNKNSKDKLSANDKNSKANDKNSAKNDKSAKLDHSDJGJASBDKN
SANDLNSMALVJOPDHVANVLNVKLSAKNFADNGKLHKNIUHSAFLSAFLFASFOLSANFLNK
FNKDSNGOIFSGHSKDHGKDHSFIABHFIHEHRFKHLKDHSUNSIUANIDAUBIOAYICMUSF
AVMAS
STFRIENDINSIDEOFADOGITSTOODARKTOREAD
3. Link the exons together
Last, the exons must be joined together to generate the final message. (The excised intron is usually broken down to its component letters, which are then recycled.)OUTSIDEOFADOGABOOKISMANSBESTFRIENDINSIDEOFADOGITSTOODARKTOREAD
Introduction
Introns were first discovered in protein-coding genes of adenovirus, and were subsequently identified in genes encoding transfer RNA and ribosomal RNA genes. Introns are now known to occur within a wide variety of genes throughout organisms and viruses within all of the biological kingdoms.The discovery of introns led to the Nobel Prize in Physiology or Medicine
Nobel Prize in Physiology or Medicine
The Nobel Prize in Physiology or Medicine administered by the Nobel Foundation, is awarded once a year for outstanding discoveries in the field of life science and medicine. It is one of five Nobel Prizes established in 1895 by Swedish chemist Alfred Nobel, the inventor of dynamite, in his will...
in 1993 for Phillip Allen Sharp
Phillip Allen Sharp
Phillip Allen Sharp is an American geneticist and molecular biologist who co-discovered RNA splicing. He shared the 1993 Nobel Prize in Physiology or Medicine with Richard J...
and Richard J. Roberts
Richard J. Roberts
Sir Richard "Rich" John Roberts is a British biochemist and molecular biologist. He was awarded the 1993 Nobel Prize in Physiology or Medicine with Phillip Allen Sharp for the discovery of introns in eukaryotic DNA and the mechanism of gene-splicing.When he was 4, his family moved to Bath. In...
. The term intron was introduced by American biochemist
Biochemistry
Biochemistry, sometimes called biological chemistry, is the study of chemical processes in living organisms, including, but not limited to, living matter. Biochemistry governs all living organisms and living processes...
Walter Gilbert
Walter Gilbert
Walter Gilbert is an American physicist, biochemist, molecular biology pioneer, and Nobel laureate.-Biography:Gilbert was born in Boston, Massachusetts, on March 21, 1932...
:
"The notion of the cistron [...] must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger - which I suggest we call introns (for intragenic regions) - alternating with regions which will be expressed - exons." (Gilbert 1978)
The frequency of introns within different genomes is observed to vary widely across the spectrum of biological organisms. For example, introns are extremely common within the nuclear genome of higher vertebrates (e.g. humans and mice), where protein-coding genes almost always contain multiple introns, while introns are rare within the nuclear genes of some eukaryotic microorganisms, for example baker's yeast
Baker's yeast
Baker's yeast is the common name for the strains of yeast commonly used as a leavening agent in baking bread and bakery products, where it converts the fermentable sugars present in the dough into carbon dioxide and ethanol...
(Saccharomyces cerevisiae). In contrast, the mitochondrial genomes
Mitochondrial DNA
Mitochondrial DNA is the DNA located in organelles called mitochondria, structures within eukaryotic cells that convert the chemical energy from food into a form that cells can use, adenosine triphosphate...
of vertebrates are entirely devoid of introns, while those of eukaryotic microorganisms may contain many introns. Introns are well known in bacterial and archaeal genes, but occur more rarely than in most eukaryotic genomes.
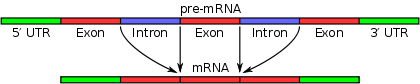
Classification
Splicing of all intron-containing RNA molecules is superficially similar, as described above. However, different types of introns were identified through the examination of intron structure by DNA sequence analysis, together with genetic and biochemical analysis of RNA splicing reactions.At least four distinct classes of introns have been identified.
- Introns in nuclear protein-coding genes that are removed by spliceosomeSpliceosomeA spliceosome is a complex of snRNA and protein subunits that removes introns from a transcribed pre-mRNA segment. This process is generally referred to as splicing.-Composition:...
s - Introns in nuclear and archaeal transfer RNA genes that are removed by proteins (tRNA splicing enzymes)
- Self-splicing group I intronsGroup I catalytic intronGroup I introns are large self-splicing ribozymes. They catalyze their own excision from mRNA, tRNA and rRNA precursors in a wide range of organisms. The core secondary structure consists of nine paired regions...
that are removed by RNA catalysisRibozymeA ribozyme is an RNA molecule with a well defined tertiary structure that enables it to catalyze a chemical reaction. Ribozyme means ribonucleic acid enzyme. It may also be called an RNA enzyme or catalytic RNA. Many natural ribozymes catalyze either the hydrolysis of one of their own...
. - Self-splicing group II intronGroup II intronGroup II introns are a large class of self-catalytic ribozymes as well as mobile genetic element found within the genes of all three domains of life. Ribozyme activity can occur under high-salt conditions in vitro. However, assistance from proteins is required for in vivo splicing...
s that are removed by RNA catalysisRibozymeA ribozyme is an RNA molecule with a well defined tertiary structure that enables it to catalyze a chemical reaction. Ribozyme means ribonucleic acid enzyme. It may also be called an RNA enzyme or catalytic RNA. Many natural ribozymes catalyze either the hydrolysis of one of their own...
Group III intron
Group III intron
Group III intron is a class of introns found in mRNA genes of chloroplasts in euglenoid protists. They have a conventional group II-type dVI with a bulged adenosine, a streamlined dI, no dII-dV, and a relaxed splice site consensus. Splicing is by two transesterification reactions with a dVI bulged...
s are proposed to be a fifth family, but little is known about the biochemical apparatus that mediates their splicing. They appear to be related to group II introns, and possibly to spliceosomal introns.
Nuclear pre-mRNA introns (spliceosomal introns) are characterized by specific intron sequences located at the boundaries between introns and exons. These sequences are recognized by spliceosomal RNA molecules when the splicing reactions are initiated. In addition, they contain a branch point, a particular nucleotide sequence near the 3' end of the intron that becomes covalently linked to the 5' end of the intron during the splicing process, generating a branched (lariat) intron. Apart from these three short conserved elements, nuclear pre-mRNA intron sequences are highly variable. Nuclear pre-mRNA introns are often much longer than their surrounding exons.
Group I and group II introns are found in genes encoding proteins (messenger RNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
), transfer RNA
Transfer RNA
Transfer RNA is an adaptor molecule composed of RNA, typically 73 to 93 nucleotides in length, that is used in biology to bridge the three-letter genetic code in messenger RNA with the twenty-letter code of amino acids in proteins. The role of tRNA as an adaptor is best understood by...
and ribosomal RNA
Ribosomal RNA
Ribosomal ribonucleic acid is the RNA component of the ribosome, the enzyme that is the site of protein synthesis in all living cells. Ribosomal RNA provides a mechanism for decoding mRNA into amino acids and interacts with tRNAs during translation by providing peptidyl transferase activity...
in a very wide range of living organisms., Following transcription into RNA, group I and group II introns also make extensive internal interactions that allow them to fold into a specific, complex three-dimensional architecture
Nucleic acid tertiary structure
300px|thumb|upright|alt = Colored dice with checkered background|Example of a large catalytic RNA. The self-splicing group II intron from Oceanobacillus iheyensis....
. These complex architectures allow some group I and group II introns to be self-splicing, that is, the intron-containing RNA molecule can rearrange its own covalent structure so as to precisely remove the intron and link the exons together in the correct order. In some cases, particular intron-binding proteins are involved in splicing, acting in such a way that the assist the intron in folding into the three-dimensional structure that is necessary for self-splicing activity. Group I and group II introns are distinguished by different sets of internal conserved sequences and folded structures, and by the fact that splicing of RNA molecules containing group II introns generates branched introns (like those of spliceosomal RNAs), while group I introns use a non-encoded guanosine nucleotide (typically GTP) to initiate splicing, adding it on to the 5'-end of the excised intron.
Transfer RNA introns that depend upon proteins for removal occur at a specific location within the anticodon loop of unspliced tRNA precursors, and are removed by a tRNA splicing endonuclease. The exons are then linked together by a second protein, the tRNA splicing ligase. Note that self-splicing introns are also sometimes found within tRNA genes.
Biological functions and evolution
As a first approximation, it is possible to view introns as unimportant sequences whose only function is to be removed from an unspliced precursor RNA in order to generate the functional mRNA, rRNA or tRNA product. However, it is now well-established that some introns themselves encode specific proteins or can be further processed after splicing to generate noncoding RNA molecules. Alternative splicingAlternative splicing
Alternative splicing is a process by which the exons of the RNA produced by transcription of a gene are reconnected in multiple ways during RNA splicing...
is widely used to generate multiple proteins from a single gene. Furthermore, some introns represent mobile genetic elements
Mobile genetic elements
Mobile genetic elements are a type of DNA that can move around within the genome. They include:*Transposons **Retrotransposons**DNA transposons**Insertion sequences*Plasmids...
and may be regarded as examples of selfish DNA
Selfish DNA
Selfish DNA refers to those sequences of DNA which, in their purest form, have two distinct properties: the DNA sequence spreads by forming additional copies of itself within the genome; and it makes no specific contribution to the reproductive success of its host organism.This idea was sketched...
.
The biological origins of introns are obscure. After the initial discovery of introns in protein-coding genes of the eukaryotic nucleus, there was significant debate as to whether introns in modern-day organisms were inherited from a common ancient ancestor (termed the introns-early hypothesis), or whether they appeared in genes rather recently in the evolutionary process (termed the introns-late hypothesis). Another theory is that the spliceosome
Spliceosome
A spliceosome is a complex of snRNA and protein subunits that removes introns from a transcribed pre-mRNA segment. This process is generally referred to as splicing.-Composition:...
and the intron-exon structure of genes is a relic of the RNA world (the introns-first hypothesis). There is still considerable debate about the extent to which of these hypotheses is most correct. The popular consensus at the moment is that introns arose within the eukaryote lineage as selfish elements.
Early studies of genomic DNA sequences from a wide range of organisms show that the intron-exon structure of homologous genes in different organisms can vary widely. More recent studies of entire eukaryotic genomes have now shown that the lengths and density (introns/gene) of introns varies considerably between related species. For example, while the human genome contains an average of 8.4 introns/gene (139,418 in the genome), the unicellular fungus Encephalitozoon cuniculi
Encephalitozoon cuniculi
Encephalitozoon cuniculi is a eukaryotic organism belonging to the phylum Microsporidia, in the kingdom Fungi. An obligate intracellular parasite, E. cuniculi occurs in laboratory mice and rabbits, monkeys, dogs, rats, birds, guinea pigs, and other mammals including humans. At various early times...
contains only 0.0075 introns/gene (15 introns in the genome). Since eukaryotes arose from a common ancestor (Common descent
Common descent
In evolutionary biology, a group of organisms share common descent if they have a common ancestor. There is strong quantitative support for the theory that all living organisms on Earth are descended from a common ancestor....
), there must have been extensive gain and/or loss of introns during evolutionary time. This process is thought to be subject to selection, with a tendency towards intron gain in larger species due to their smaller population sizes, and the converse in smaller (particularly unicellular) species. Biological factors also influence which genes in a genome lose or accumulate introns.
Alternative splicing
Alternative splicing
Alternative splicing is a process by which the exons of the RNA produced by transcription of a gene are reconnected in multiple ways during RNA splicing...
of introns within a gene acts to introduce greater variability of protein sequences translated from a single gene, allowing multiple related proteins to be generated from a single gene and a single precursor mRNA transcript. The control of alternative RNA splicing is performed by complex network of signaling molecules that respond to a wide range of intracellular and extracellular signals.
Introns contain several short sequences that are important for efficient splicing, such as acceptor and donor sites at either end of the intron as well as a branch point site, which are required for proper splicing by the spliceosome
Spliceosome
A spliceosome is a complex of snRNA and protein subunits that removes introns from a transcribed pre-mRNA segment. This process is generally referred to as splicing.-Composition:...
. Some introns are known to enhance the expression of the gene that they are contained in by a process known as intron-mediated enhancement (IME).
See also
Structure:- ExonExonAn exon is a nucleic acid sequence that is represented in the mature form of an RNA molecule either after portions of a precursor RNA have been removed by cis-splicing or when two or more precursor RNA molecules have been ligated by trans-splicing. The mature RNA molecule can be a messenger RNA...
- mRNA
- Eukaryotic chromosome fine structureEukaryotic chromosome fine structureEukaryotic chromosome fine structure refers to the structure of sequences for eukaryotic chromosomes. Some fine sequences are included in more than one class, so the classification listed is not intended to be completely separate.-Chromosomal characteristics:...
Splicing:
- Alternative splicingAlternative splicingAlternative splicing is a process by which the exons of the RNA produced by transcription of a gene are reconnected in multiple ways during RNA splicing...
- Minor spliceosomeMinor spliceosomeThe minor spliceosome is a ribonucleoprotein complex that catalyses the removal of an atypical class of spliceosomal introns from eukaryotic messenger RNAs in plant, insects, vertebrates and some fungi . This process is called noncanonical splicing, as opposed to U2-dependent canonical splicing...
Function
- MicroRNA
Others:
- InteinInteinAn intein is a segment of a protein that is able to excise itself and rejoin the remaining portions with a peptide bond. Inteins have also been called "protein introns"....
- Interrupted geneInterrupted geneAn interrupted gene is simply a strand of DNA that contains both introns and exons. Most higher-level eukaryotes have interrupted genes and have longer introns than exons, creating a gene that is longer than its coding region. Interrupted genes are also found in some bacteria...
- Noncoding DNANoncoding DNAIn genetics, noncoding DNA describes components of an organism's DNA sequences that do not encode for protein sequences. In many eukaryotes, a large percentage of an organism's total genome size is noncoding DNA, although the amount of noncoding DNA, and the proportion of coding versus noncoding...
- Noncoding RNA
- Selfish DNASelfish DNASelfish DNA refers to those sequences of DNA which, in their purest form, have two distinct properties: the DNA sequence spreads by forming additional copies of itself within the genome; and it makes no specific contribution to the reproductive success of its host organism.This idea was sketched...
- TwintronTwintronTwintrons are introns-within-introns excised by sequential splicing reactions. Twintrons are presumably formed by the insertion of a mobile intron into an existing intron....
External links
- A search engine for exon/intron sequences defined by NCBI
- Bruce Alberts, Alexander Johnson, Julian Lewis, Martin Raff, Keith Roberts, and Peter Walter Molecular Biology of the Cell, 2007, ISBN 978-0-8153-4105-5. Fourth edition is available online through the NCBI Bookshelf: link
- Jeremy M Berg, John L Tymoczko, and Lubert Stryer, Biochemistry 5th edition, 2002, W H Freeman. Available online through the NCBI Bookshelf: link
- Intron finding tool for plant genomic sequences
- Exon-intron graphic maker

