
Exon
Encyclopedia
An exon is a nucleic acid
sequence that is represented in the mature form of an RNA
molecule either after portions of a precursor RNA (intron
s) have been removed by cis-splicing
or when two or more precursor RNA molecules have been ligated
by trans-splicing
. The mature RNA molecule can be a messenger RNA
or a functional form of a non-coding RNA
such as rRNA or tRNA. Depending on the context, exon can refer to the sequence in the DNA or its RNA transcript.
Walter Gilbert
in 1978: "The notion of the cistron
… must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger which I suggest we call introns (for intragenic regions) alternating with regions which will be expressed exons."
This definition was originally made for protein-coding transcripts that are spliced before being translated. The term later came to include sequences removed from rRNA and tRNA, and it also was used later for RNA molecules originating from different parts of the genome that are then ligated by trans-splicing.
, each of the exons contains part of the open reading frame
(ORF) that codes for a specific portion of the complete protein
. However, the term exon is often misused to refer only to coding sequences for the final protein. This is incorrect, since many noncoding exons are known in human genes (Zhang 1998).
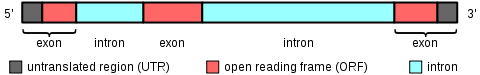
To the right is a diagram of a heterogeneous nuclear RNA (hnRNA), which is an unedited mRNA transcript, or pre-mRNAs. Exons can include both sequences that code for amino acids (red) and untranslated sequences (grey). Stretches of unused sequence called intron
s (blue) are removed, and the exons are joined together to form the final functional mRNA
. The notation 5' and 3' refer to the direction of the DNA template in the chromosome and is used to distinguish between the two untranslated regions (grey).
Some of the exons will be wholly or part of the 5' untranslated region (5' UTR
) or the 3' untranslated region (3' UTR
) of each transcript. The untranslated regions are important for efficient translation of the transcript
and for controlling the rate of translation and half-life of the transcript. Furthermore, transcripts made from the same gene may not have the same exon structure, since parts of the mRNA could be removed by the process of alternative splicing
. Some mRNA transcripts have exons with no ORFs and, thus, are sometimes referred to as non-coding RNA
.
Exonization is the creation of a new exon, as result of mutations in intron
ic sequences.
Polycistronic messages have multiple ORFs in one transcript and also have small regions of untranslated sequence between each ORF.
or 'gene trapping
' is a molecular biology
technique that exploits the existence of the intron-exon splicing
to find new genes. The first exon of a 'trapped' gene splices into the exon that is contained in the insertional DNA. This new exon contains the ORF for a reporter gene
that can now be expressed using the enhancer
s that control the target gene. A scientist knows that a new gene has been trapped when the reporter gene is expressed.
Splicing can be experimentally modified so that targeted exons are excluded from mature mRNA transcripts by blocking the access of splice-directing small nuclear ribonucleoprotein particles (snRNPs) to pre-mRNA using Morpholino antisense oligos
. This has become a standard technique in developmental biology
. Morpholino oligos can also be targeted to prevent molecules that regulate splicing (e.g. splice enhancers, splice suppressors) from binding to pre-mRNA, altering patterns of splicing.
Nucleic acid
Nucleic acids are biological molecules essential for life, and include DNA and RNA . Together with proteins, nucleic acids make up the most important macromolecules; each is found in abundance in all living things, where they function in encoding, transmitting and expressing genetic information...
sequence that is represented in the mature form of an RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
molecule either after portions of a precursor RNA (intron
Intron
An intron is any nucleotide sequence within a gene that is removed by RNA splicing to generate the final mature RNA product of a gene. The term intron refers to both the DNA sequence within a gene, and the corresponding sequence in RNA transcripts. Sequences that are joined together in the final...
s) have been removed by cis-splicing
RNA splicing
In molecular biology and genetics, splicing is a modification of an RNA after transcription, in which introns are removed and exons are joined. This is needed for the typical eukaryotic messenger RNA before it can be used to produce a correct protein through translation...
or when two or more precursor RNA molecules have been ligated
Ligase
In biochemistry, ligase is an enzyme that can catalyse the joining of two large molecules by forming a new chemical bond, usually with accompanying hydrolysis of a small chemical group dependent to one of the larger molecules...
by trans-splicing
Trans-splicing
Trans-splicing is a special form of RNA processing in eukaryotes where exons from two different primary RNA transcripts are joined end to end and ligated....
. The mature RNA molecule can be a messenger RNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
or a functional form of a non-coding RNA
Non-coding RNA
A non-coding RNA is a functional RNA molecule that is not translated into a protein. Less-frequently used synonyms are non-protein-coding RNA , non-messenger RNA and functional RNA . The term small RNA is often used for short bacterial ncRNAs...
such as rRNA or tRNA. Depending on the context, exon can refer to the sequence in the DNA or its RNA transcript.
History
The term exon derives from expressed region and was coined by American biochemistBiochemistry
Biochemistry, sometimes called biological chemistry, is the study of chemical processes in living organisms, including, but not limited to, living matter. Biochemistry governs all living organisms and living processes...
Walter Gilbert
Walter Gilbert
Walter Gilbert is an American physicist, biochemist, molecular biology pioneer, and Nobel laureate.-Biography:Gilbert was born in Boston, Massachusetts, on March 21, 1932...
in 1978: "The notion of the cistron
Cistron
A cistron is a gene. The term cistron is used to emphasize that genes exhibit a specific behavior in a cis-trans test; distinct positions within a genome are cistronic when mutations at the loci exhibit the same simple Mendelian inheritance as would mutations at a single locus.For example,...
… must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger which I suggest we call introns (for intragenic regions) alternating with regions which will be expressed exons."
This definition was originally made for protein-coding transcripts that are spliced before being translated. The term later came to include sequences removed from rRNA and tRNA, and it also was used later for RNA molecules originating from different parts of the genome that are then ligated by trans-splicing.
Function
In many genesGênes
Gênes is the name of a département of the First French Empire in present Italy, named after the city of Genoa. It was formed in 1805, when Napoleon Bonaparte occupied the Republic of Genoa. Its capital was Genoa, and it was divided in the arrondissements of Genoa, Bobbio, Novi Ligure, Tortona and...
, each of the exons contains part of the open reading frame
Open reading frame
In molecular genetics, an open reading frame is a DNA sequence that does not contain a stop codon in a given reading frame.Normally, inserts which interrupt the reading frame of a subsequent region after the start codon cause frameshift mutation of the sequence and dislocate the sequences for stop...
(ORF) that codes for a specific portion of the complete protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
. However, the term exon is often misused to refer only to coding sequences for the final protein. This is incorrect, since many noncoding exons are known in human genes (Zhang 1998).

To the right is a diagram of a heterogeneous nuclear RNA (hnRNA), which is an unedited mRNA transcript, or pre-mRNAs. Exons can include both sequences that code for amino acids (red) and untranslated sequences (grey). Stretches of unused sequence called intron
Intron
An intron is any nucleotide sequence within a gene that is removed by RNA splicing to generate the final mature RNA product of a gene. The term intron refers to both the DNA sequence within a gene, and the corresponding sequence in RNA transcripts. Sequences that are joined together in the final...
s (blue) are removed, and the exons are joined together to form the final functional mRNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
. The notation 5' and 3' refer to the direction of the DNA template in the chromosome and is used to distinguish between the two untranslated regions (grey).
Some of the exons will be wholly or part of the 5' untranslated region (5' UTR
Five prime untranslated region
A messenger ribonucleic acid molecule codes for a protein through translation. The mRNA also contains regions that are not translated: in eukaryotes these include the 5' untranslated region, 3' untranslated region, 5' cap and poly-A tail....
) or the 3' untranslated region (3' UTR
Three prime untranslated region
In molecular genetics, the three prime untranslated region is a particular section of messenger RNA . It is preceeded by the coding region....
) of each transcript. The untranslated regions are important for efficient translation of the transcript
Transcript
Transcript may refer to:* Transcript , a copy of a student's permanent academic record* Transcription , the process of creating an equivalent RNA copy of a sequence of DNA* Transcript , a record of all court proceedings...
and for controlling the rate of translation and half-life of the transcript. Furthermore, transcripts made from the same gene may not have the same exon structure, since parts of the mRNA could be removed by the process of alternative splicing
Alternative splicing
Alternative splicing is a process by which the exons of the RNA produced by transcription of a gene are reconnected in multiple ways during RNA splicing...
. Some mRNA transcripts have exons with no ORFs and, thus, are sometimes referred to as non-coding RNA
Non-coding RNA
A non-coding RNA is a functional RNA molecule that is not translated into a protein. Less-frequently used synonyms are non-protein-coding RNA , non-messenger RNA and functional RNA . The term small RNA is often used for short bacterial ncRNAs...
.
Exonization is the creation of a new exon, as result of mutations in intron
Intron
An intron is any nucleotide sequence within a gene that is removed by RNA splicing to generate the final mature RNA product of a gene. The term intron refers to both the DNA sequence within a gene, and the corresponding sequence in RNA transcripts. Sequences that are joined together in the final...
ic sequences.
Polycistronic messages have multiple ORFs in one transcript and also have small regions of untranslated sequence between each ORF.
Experimental approaches that utilize exons
Exon trappingExon trapping
Exon trapping is a molecular biology technique to identify potential exons in a fragment of eukaryote DNA of unknown intron-exon structure. This is done to determine if the fragment is part of an expressed gene....
or 'gene trapping
Gene trapping
Gene trapping is a high-throughput approach that is used to introduce insertional mutations across the mammalian genome. It is performed with gene trap vectors whose principal element is a gene trapping cassette consisting of a promoterless reporter gene and/or selectable genetic marker flanked by...
' is a molecular biology
Molecular biology
Molecular biology is the branch of biology that deals with the molecular basis of biological activity. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry...
technique that exploits the existence of the intron-exon splicing
RNA splicing
In molecular biology and genetics, splicing is a modification of an RNA after transcription, in which introns are removed and exons are joined. This is needed for the typical eukaryotic messenger RNA before it can be used to produce a correct protein through translation...
to find new genes. The first exon of a 'trapped' gene splices into the exon that is contained in the insertional DNA. This new exon contains the ORF for a reporter gene
Reporter gene
In molecular biology, a reporter gene is a gene that researchers attach to a regulatory sequence of another gene of interest in cell culture, animals or plants. Certain genes are chosen as reporters because the characteristics they confer on organisms expressing them are easily identified and...
that can now be expressed using the enhancer
Enhancer (genetics)
In genetics, an enhancer is a short region of DNA that can be bound with proteins to enhance transcription levels of genes in a gene cluster...
s that control the target gene. A scientist knows that a new gene has been trapped when the reporter gene is expressed.
Splicing can be experimentally modified so that targeted exons are excluded from mature mRNA transcripts by blocking the access of splice-directing small nuclear ribonucleoprotein particles (snRNPs) to pre-mRNA using Morpholino antisense oligos
Morpholino
In molecular biology, a Morpholino is a molecule in a particular structural family that is used to modify gene expression. Morpholino oligomers are an antisense technology used to block access of other molecules to specific sequences within nucleic acid...
. This has become a standard technique in developmental biology
Developmental biology
Developmental biology is the study of the process by which organisms grow and develop. Modern developmental biology studies the genetic control of cell growth, differentiation and "morphogenesis", which is the process that gives rise to tissues, organs and anatomy.- Related fields of study...
. Morpholino oligos can also be targeted to prevent molecules that regulate splicing (e.g. splice enhancers, splice suppressors) from binding to pre-mRNA, altering patterns of splicing.
See also
- Eukaryotic gene example
- Exon shufflingExon shufflingExon shuffling is a theory, introduced by Walter Gilbert in 1977, in which different exons either within a gene or between two nonallelic genes are occasionally mixed. Gilbert suggested that exons might each encode a single protein domain, establishing a kind of modular property...
- Interrupted geneInterrupted geneAn interrupted gene is simply a strand of DNA that contains both introns and exons. Most higher-level eukaryotes have interrupted genes and have longer introns than exons, creating a gene that is longer than its coding region. Interrupted genes are also found in some bacteria...
- IntronIntronAn intron is any nucleotide sequence within a gene that is removed by RNA splicing to generate the final mature RNA product of a gene. The term intron refers to both the DNA sequence within a gene, and the corresponding sequence in RNA transcripts. Sequences that are joined together in the final...
- mRNA
- Untranslated region (UTR)

