P-type ATPase
Encyclopedia
The P-type ATPases, also known as E1-E2 ATPases, are a large group of evolutionarily related ion pumps that are found in bacteria
, archaea
and eukaryotes. They are α-helical bundle
primary transporters
referred to as P-type ATPases because they catalyze auto- (or self-) phosphorylation
of a key conserved aspartate residue within the pump. In addition, they all appear to interconvert between at least two different conformations, denoted by E1 and E2.
Most members of this transporter family are specific for the pumping of a large array of cations, however one subfamily is involved in flipping phospholipids to maintain the asymmetric nature of the biomembrane.
Prominent examples of P-type ATPases are the sodium-potassium pump (Na+,K+-ATPase), the proton pump
(H+-ATPase), the proton-potassium pump
(H+,K+-ATPase) and the calcium pump
(Ca2+-ATPase).
isolated in 1957. The Na+,K+-ATPase was only the first member of a large and still growing protein family, which in June 2007 had around 400 confirmed and unique members in Swiss-Prot (Prosite motif PS00154).
, and eucaryota. This underlines the significance of this protein family for cell survival.
Methanococcus
jannaschii deserves special attention, because it might reveal information about the evolution of the ancestor P-type ATPase. This open reading frame
covers the big cytoplasmic loop common to all P-type ATPases, which contain all the amino acids involved in ATP binding and hydrolysis. It is speculated that this soluble protein has fused with an ion channel, thus creating the first P-type ATPase.
, a sarco(endo)plasmic reticulum Ca2+-ATPase of fast twitch muscle from adult rabbit
. It is generally acknowledged that the structure of SERCA1a
is representative for the family of P-type ATPases.
Structures of the Na+/K+-ATPase and H+-ATPase
are also available.
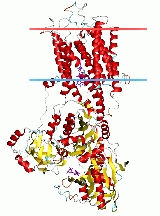
is composed of a cytoplasmic section and a transmembrane section with two Ca2+ binding sites. The cytoplasmic section consists of three cytoplasmic domains, designated the P, N and A domains, containing over half the mass of the protein. The transmembrane section has ten transmembrane helices
(M1-M10), with the two Ca2+ binding sites located near the midpoint of the bilayer. The binding sites are formed by side-chains and backbone carbonyls from M4, M5, M6, and M8. M4 is unwound in this region due to a conserved proline (P308). This unwinding of M4 is recognised as a key structural feature of P-type ATPases.
The P domain contains the canonical aspartic acid phosphorylated during the reaction cycle. It is composed of two parts widely separated in sequence. These two parts assemble into a seven stranded parallel ß-sheet with eight short associated a-helices, forming a Rossmann fold
.
The N domain is inserted between the two segments of the P domain, and is formed of a seven strand antiparallel ß-sheet between two helix bundles. This domain contains the ATP-binding pocket, pointing out toward the solvent near the P-domain.
The A domain is the smallest of the three domains. It consists of a distorted jellyroll structure and two short helices. It is the actuator domain modulating the occlusion of Ca2+ in the transmembrane binding sites, and it is pivot in transposing the energy from the hydrolysis of ATP in the cytoplasmic domains to the vectorial transport of cations in the transmembrane domain. The A domain dephosphorylates the P domain as part of the reaction cycle using a highly conserved TGES motif located at one end of the jellyroll.
ATP hydrolysis occurs in the cytoplasmic headpiece at the interface between domain N and P. Two Mg-ion sites forms part of the active site. ATP hydrolysis is tightly coupled to Ca2+ translocation through the membrane, more than 40 Å away, by the A domain.
It is interesting to note that the folding pattern and the locations of the critical amino acids for phosphorylation in P-type ATPases has the haloacid dehalogenase fold characteristic of the haloacid dehalogenase (HAD) superfamily, as predicted by sequence homology. The HAD superfamily functions on the common theme of an aspartate ester formation by an SN2 reaction
mechanism. This SN2 reaction
is clearly observed in the solved structure of SERCA with ADP
plus AlF4-.
and other P-IIA ATPases are also regulated by phospholamban and sarcolipin in vivo. Probably other subfamilies also need additional subunits for the proper function in vivo.
Some members of the family have additional domains fused to the pump. Heavy metal pumps can have several N- and C-terminal heavy metal-binding domains
that have been found to be involved in regulation.
The proton pumps (IIIA) have a C-terminal regulatory domain (called the R domain), which, when unphosphorylated, inhibit pumping.
While most subfamilies have 10 transmembrane helices, there are some notable exceptions. The P-IA ATPases are predicted to have 7, and the large subfamily of heavy metal pumps (IB) is predicted to have 8 transmembrane helices. Type V appears to have a total of 12 transmembrane helices.
The E1-E2 schema has been proven to work, but there exist more than two major conformational states. However, the E1-E2 notation highlights the selectivity of the enzyme
. In E1, the pump has high affinity for the exported substrate and low affinity for the imported substrate. In E2, it has low affinity of the exported substrate and high affinity for the imported substrate.
Four major enzyme states form the cornerstones in the reaction cycle. Several additional reaction intermediates occur interposed. These are termed E1~P, E2P, E2-P*, and E1/E2, described below.
In the case of SERCA1a
, energy from ATP
is used to transport 2 Ca2+-ions from the cytoplasmic side to the lumen
of the sarcoplasmatic reticulum, and to countertransport 1-3 protons into the cytoplasm
.
Starting in the E1/E2 state, the reaction cycle begins as the enzyme releases 1-3 protons from the cation-ligating residues, in exchange for cytoplasmic Ca2+-ions. This leads to assembly of the phosphorylation site between the ATP-bound N domain and the P domain, while the A domain directs the occlusion of the bound Ca2+. In this occluded state, the Ca2+ ions are buried in a proteinacious environment with no access to either side of the membrane.
The Ca2E1~P state becomes formed through a kinase reaction, where the P domain becomes phosphorylated, producing ADP. The cleavage of the ß,-phosphordiester bond releases the gamma-phosphate from ADP and unleashes the N domain from the P domain.
This then allows the A domain to rotate towards the phosphorylation site, making a firm association with both the P and the N domain. This movement of the A domain exerts a downward push on M3-M4 and a drag on M1-M2, forcing the pump to open at the luminal side and forming the E2P state. During this transition, the transmembrane Ca2+-binding residues are forces apart, destroying the high-affinity binding site. This is in agreement with the general model form substrate translocation (cf. 1.2), showing that energy in primary transport is not used to bind the substrate but to release it again from the buried counter ions. At the same time the N domain becomes exposed to the cytosol, ready for ATP exchange at the nucleotide-binding site.
As the Ca2+ dissociate to the luminal side, the cation binding sites are neutralised by proton binding, and this make a closure of the transmembrane segments favourable. This closure is coupled to a downwards rotation of the A domain and a movement of the P domain, which then leads to the E2-P* occluded state. Meanwhile, the N domain exchanges ADP for ATP.
The P domain is dephosphorylated by the A domain, and the cycle completes when the phosphate is released from the enzyme, stimulated by the newly bound ATP, while a cytoplasmic pathway opens to exchange the protons for two new Ca2+-ions..
Bacteria
Bacteria are a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a wide range of shapes, ranging from spheres to rods and spirals...
, archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
and eukaryotes. They are α-helical bundle
Transmembrane protein
A transmembrane protein is a protein that goes from one side of a membrane through to the other side of the membrane. Many TPs function as gateways or "loading docks" to deny or permit the transport of specific substances across the biological membrane, to get into the cell, or out of the cell as...
primary transporters
Active transport
Active transport is the movement of a substance against its concentration gradient . In all cells, this is usually concerned with accumulating high concentrations of molecules that the cell needs, such as ions, glucose, and amino acids. If the process uses chemical energy, such as from adenosine...
referred to as P-type ATPases because they catalyze auto- (or self-) phosphorylation
Phosphorylation
Phosphorylation is the addition of a phosphate group to a protein or other organic molecule. Phosphorylation activates or deactivates many protein enzymes....
of a key conserved aspartate residue within the pump. In addition, they all appear to interconvert between at least two different conformations, denoted by E1 and E2.
Most members of this transporter family are specific for the pumping of a large array of cations, however one subfamily is involved in flipping phospholipids to maintain the asymmetric nature of the biomembrane.
Prominent examples of P-type ATPases are the sodium-potassium pump (Na+,K+-ATPase), the proton pump
Proton ATPase
In the field of enzymology, the proton-ATPase is an enzyme that catalyzes the following chemical reaction:The 3 substrates of this enzyme are ATP, H2O, and H+, whereas its 3 products are ADP, phosphate, and H+....
(H+-ATPase), the proton-potassium pump
Hydrogen potassium ATPase
Gastric hydrogen potassium ATPase is also known as H+/K+ ATPase- Function and location :The gastric hydrogen potassium ATPase or H+/K+ ATPase is the proton pump of the stomach and, as such, is the enzyme primarily responsible for the acidification of the stomach contents...
(H+,K+-ATPase) and the calcium pump
SERCA
SERCA, or sarco/endoplasmic reticulum Ca2+-ATPase, or SR Ca2+-ATPase, is a calcium ATPase-type P-ATPase.-Function:SERCA resides in the sarcoplasmic reticulum within muscle cells...
(Ca2+-ATPase).
Discovery
The first P-type ATPase discovered was the Na+,K+-ATPase, which Nobel laureate Jens Christian SkouJens Christian Skou
Jens Christian Skou is a Danish chemist and Nobel laureate.Skou was born in Lemvig, Denmark to a wealthy family. His father Magnus Martinus Skou was a timber and coal merchant. His mother Ane-Margrethe Skou took over the company after the death of his father. At the age of 15 Skou entered a...
isolated in 1957. The Na+,K+-ATPase was only the first member of a large and still growing protein family, which in June 2007 had around 400 confirmed and unique members in Swiss-Prot (Prosite motif PS00154).
Phylogenetic classification
A phylogenetic analysis of 159 sequences made in 1998 by Axelsen and Palmgren showed that P-type ATPases can be divided into five subfamilies, based strictly on a conserved sequence kernel excluding the highly variable N and C terminal regions. The phylogenetic analysis grouped the proteins independent of the organism from which they are isolated and showed that the diversification of the P-type ATPase family occurred prior to the separation of eubacteria, archaeaArchaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
, and eucaryota. This underlines the significance of this protein family for cell survival.
- Type I consists of the transition/heavy metal ATPases.
- Type IA ATPases are involved in K+ import. They are atypical P-type ATPases because, unlike other P-type ATPases, they function as part of a heterotetrameric complex (called KdpFABC), where the actual K+ transport is mediated by another subcomponent of the complex.
- Type IB ATPases are involved in transport of the soft Lewis acidsLewis acid]The term Lewis acid refers to a definition of acid published by Gilbert N. Lewis in 1923, specifically: An acid substance is one which can employ a lone pair from another molecule in completing the stable group of one of its own atoms. Thus, H+ is a Lewis acid, since it can accept a lone pair,...
: Cu+, Ag+, Cu2+, Zn2+, Cd2+, Pb2+ and Co2+. They are key elements for metal resistance and metal homeostasis in a wide range of organisms.
- Type II ATPases are split into four groups.
- Type IIA transports Ca2+. SERCA1aSERCASERCA, or sarco/endoplasmic reticulum Ca2+-ATPase, or SR Ca2+-ATPase, is a calcium ATPase-type P-ATPase.-Function:SERCA resides in the sarcoplasmic reticulum within muscle cells...
is a type IIA pump. - Type IIB transports Ca2+.
- Type IIC consists of the closely related Na+/K+ and H+/K+ ATPases from animal cells.
- Type IID contains a small number of fungal ATPases of unknown function.
- Type IIA transports Ca2+. SERCA1a
- Type III ATPases contains the plasma membrane H+-ATPases from plants and fungi (IIIA) and a small subdivision with Mg2+-ATPases from three bacterial species (IIIB).
- Type IV ATPases have been shown to be involved in the transport of phospholipids. However the transport specificity of the P-IV type ATPases still remains a somewhat controversial subject.
- Type V ATPases have unknown specificity. This large group are only found in eukaryotes and are believed to be involved in cation transport in the endoplasmic reticulumEndoplasmic reticulumThe endoplasmic reticulum is an organelle of cells in eukaryotic organisms that forms an interconnected network of tubules, vesicles, and cisternae...
.
Human genes
Human genes encoding P-type ATPases or P-type ATPase-like proteins include:- Na+/K+ transportingNa+/K+-ATPaseNa+/K+-ATPase is an enzyme located in the plasma membrane in all animals.- Sodium-potassium pumps :Active transport is responsible for cells containing relatively high...
: ATP1A1, ATP1A2ATP1A2ATPase, Na+/K+ transporting, alpha 2 polypeptide, also known as ATP1A2, is a protein which in humans is encoded by the ATP1A2 gene.- Function :...
, ATP1A3ATP1A3Sodium/potassium-transporting ATPase subunit alpha-3 is an enzyme that in humans is encoded by the ATP1A3 gene.The protein encoded by this gene belongs to the family of P-type cation transport ATPases, and to the subfamily of Na+/K+-ATPases...
, ATP1A4ATP1A4Sodium/potassium-transporting ATPase subunit alpha-4 is an enzyme that in humans is encoded by the ATP1A4 gene.-Further reading:...
, ATP1B1ATP1B1Sodium/potassium-transporting ATPase subunit beta-1 is an enzyme that in humans is encoded by the ATP1B1 gene.-Further reading:...
, ATP1B2, ATP1B3ATP1B3Sodium/potassium-transporting ATPase subunit beta-3 is an enzyme that in humans is encoded by the ATP1B3 gene. ATP1B3 has also been designated as CD298 .-External links:...
, ATP1B4 - Ca++ transportingCalcium ATPaseCalcium ATPase is a form of P-ATPase that transfers calcium after a muscle has contracted. The calcium ATPase are:*Plasma membrane Ca2+ ATPase *Sarcoplasmic reticulum Ca2+ ATPase - Plasma membrane Ca2+ ATPase :...
: ATP2A1ATP2A1Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 is an enzyme that in humans is encoded by the ATP2A1 gene.-Interactions:ATP2A1 has been shown to interact with PLN and SLN.-Further reading:...
, ATP2A2ATP2A2ATP2A2 is an ATPase associated with Darier's disease and Acrokeratosis verruciformis.This gene encodes one of the SERCA Ca-ATPases, which are intracellular pumps located in the sarcoplasmic or endoplasmic reticula of muscle cells...
, ATP2A3ATP2A3Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 is an enzyme that in humans is encoded by the ATP2A3 gene.-Further reading:...
, ATP2B1ATP2B1Plasma membrane calcium-transporting ATPase 1 is an enzyme that in humans is encoded by the ATP2B1 gene.-Further reading:...
, ATP2B2ATP2B2Plasma membrane calcium-transporting ATPase 2 is an enzyme that in humans is encoded by the ATP2B2 gene.-Further reading:...
, ATP2B3ATP2B3Plasma membrane calcium-transporting ATPase 3 is an enzyme that in humans is encoded by the ATP2B3 gene.-Further reading:...
, ATP2B4ATP2B4Plasma membrane calcium-transporting ATPase 4 is an enzyme that in humans is encoded by the ATP2B4 gene.-Further reading:...
, ATP2C1ATP2C1Calcium-transporting ATPase type 2C member 1 is an enzyme that in humans is encoded by the ATP2C1 gene.-Further reading:... - Cu++ transporting: ATP7AATP7ACopper-transporting ATPase 1 also known as copper pump 1 or Menkes disease-associated protein is a protein that in humans is encoded by the ATP7A gene.- Gene :...
, ATP7B - Class I, type 8: ATP8A1, ATP8B1ATP8B1Probable phospholipid-transporting ATPase IC is an enzyme that in humans is encoded by the ATP8B1 gene. This protein is associated with progressive familial intrahepatic cholestasis type 1 as well as benign recurrent intrahepatic cholestasis.- Function :...
, ATP8B2, ATP8B3ATP8B3ATPase, aminophospholipid transporter, class I, type 8B, member 3 is a protein that in humans is encoded by the ATP8B3 gene.The protein encoded by this gene belongs to the family of P-type cation transport ATPases, and to the subfamily of aminophospholipid-transporting ATPases...
, ATP8B4 - Class II, type 9: ATP9A, ATP9B
- Class V, type 10: ATP10AATP10AProbable phospholipid-transporting ATPase VA also known as ATPase class V type 10A or aminophospholipid translocase VA is an enzyme that in humans is encoded by the ATP10A gene.- Function :...
, ATP10B, ATP10D - Class VI, type 11: ATP11A, ATP11BATP11BProbable phospholipid-transporting ATPase IF is an enzyme that in humans is encoded by the ATP11B gene.-Further reading:...
, ATP11C - H+/K+ transporting, nongastric: ATP12AATP12AATPase, H+/K+ transporting, nongastric, alpha polypeptide is a protein that in humans is encoded by the ATP12A gene.- Function :The protein encoded by this gene belongs to the family of P-type cation transport ATPases...
- type 13: ATP13A1, ATP13A2ATP13A2Probable cation-transporting ATPase 13A2 is an enzyme that in humans is encoded by the ATP13A2 gene.-Further reading:...
, ATP13A3ATP13A3Probable cation-transporting ATPase 13A3 is an enzyme that in humans is encoded by the ATP13A3 gene.-Further reading:...
, ATP13A4, ATP13A5 - ATP2C2;
- ATP4A;
- KIAA0195
Evolutionary origin
One of the open reading frames in the genome of the archaeaArchaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
Methanococcus
Methanococcus
In taxonomy, Methanococcus is a genus of the Methanococcaceae.Methanococcus is a genus of coccoid methanogens. They are all mesophiles, except the thermophilic M. thermolithotrophicus and the hyperthermophilic M. jannaschii...
jannaschii deserves special attention, because it might reveal information about the evolution of the ancestor P-type ATPase. This open reading frame
Open reading frame
In molecular genetics, an open reading frame is a DNA sequence that does not contain a stop codon in a given reading frame.Normally, inserts which interrupt the reading frame of a subsequent region after the start codon cause frameshift mutation of the sequence and dislocate the sequences for stop...
covers the big cytoplasmic loop common to all P-type ATPases, which contain all the amino acids involved in ATP binding and hydrolysis. It is speculated that this soluble protein has fused with an ion channel, thus creating the first P-type ATPase.
Structure
Most of our knowledge about the structure and function of P-type ATPases originates from SERCA1aSERCA
SERCA, or sarco/endoplasmic reticulum Ca2+-ATPase, or SR Ca2+-ATPase, is a calcium ATPase-type P-ATPase.-Function:SERCA resides in the sarcoplasmic reticulum within muscle cells...
, a sarco(endo)plasmic reticulum Ca2+-ATPase of fast twitch muscle from adult rabbit
Rabbit
Rabbits are small mammals in the family Leporidae of the order Lagomorpha, found in several parts of the world...
. It is generally acknowledged that the structure of SERCA1a
SERCA
SERCA, or sarco/endoplasmic reticulum Ca2+-ATPase, or SR Ca2+-ATPase, is a calcium ATPase-type P-ATPase.-Function:SERCA resides in the sarcoplasmic reticulum within muscle cells...
is representative for the family of P-type ATPases.
Structures of the Na+/K+-ATPase and H+-ATPase
Proton ATPase
In the field of enzymology, the proton-ATPase is an enzyme that catalyzes the following chemical reaction:The 3 substrates of this enzyme are ATP, H2O, and H+, whereas its 3 products are ADP, phosphate, and H+....
are also available.
Structure of SERCA1a
SERCA1aSERCA
SERCA, or sarco/endoplasmic reticulum Ca2+-ATPase, or SR Ca2+-ATPase, is a calcium ATPase-type P-ATPase.-Function:SERCA resides in the sarcoplasmic reticulum within muscle cells...
is composed of a cytoplasmic section and a transmembrane section with two Ca2+ binding sites. The cytoplasmic section consists of three cytoplasmic domains, designated the P, N and A domains, containing over half the mass of the protein. The transmembrane section has ten transmembrane helices
Transmembrane protein
A transmembrane protein is a protein that goes from one side of a membrane through to the other side of the membrane. Many TPs function as gateways or "loading docks" to deny or permit the transport of specific substances across the biological membrane, to get into the cell, or out of the cell as...
(M1-M10), with the two Ca2+ binding sites located near the midpoint of the bilayer. The binding sites are formed by side-chains and backbone carbonyls from M4, M5, M6, and M8. M4 is unwound in this region due to a conserved proline (P308). This unwinding of M4 is recognised as a key structural feature of P-type ATPases.
The P domain contains the canonical aspartic acid phosphorylated during the reaction cycle. It is composed of two parts widely separated in sequence. These two parts assemble into a seven stranded parallel ß-sheet with eight short associated a-helices, forming a Rossmann fold
Rossmann fold
The Rossmann fold is a protein structural motif found in proteins that bind nucleotides, especially the cofactor NAD. The structure with two repeats is composed of six parallel beta strands linked to two pairs of alpha helices in the topological order beta-alpha-beta-alpha-beta...
.
The N domain is inserted between the two segments of the P domain, and is formed of a seven strand antiparallel ß-sheet between two helix bundles. This domain contains the ATP-binding pocket, pointing out toward the solvent near the P-domain.
The A domain is the smallest of the three domains. It consists of a distorted jellyroll structure and two short helices. It is the actuator domain modulating the occlusion of Ca2+ in the transmembrane binding sites, and it is pivot in transposing the energy from the hydrolysis of ATP in the cytoplasmic domains to the vectorial transport of cations in the transmembrane domain. The A domain dephosphorylates the P domain as part of the reaction cycle using a highly conserved TGES motif located at one end of the jellyroll.
ATP hydrolysis occurs in the cytoplasmic headpiece at the interface between domain N and P. Two Mg-ion sites forms part of the active site. ATP hydrolysis is tightly coupled to Ca2+ translocation through the membrane, more than 40 Å away, by the A domain.
It is interesting to note that the folding pattern and the locations of the critical amino acids for phosphorylation in P-type ATPases has the haloacid dehalogenase fold characteristic of the haloacid dehalogenase (HAD) superfamily, as predicted by sequence homology. The HAD superfamily functions on the common theme of an aspartate ester formation by an SN2 reaction
SN2 reaction
The SN2 reaction is a type of nucleophilic substitution, where a lone pair from a nucleophile attacks an electron deficient electrophilic center and bonds to it, expelling another group called a leaving group. Thus the incoming group replaces the leaving group in one step...
mechanism. This SN2 reaction
SN2 reaction
The SN2 reaction is a type of nucleophilic substitution, where a lone pair from a nucleophile attacks an electron deficient electrophilic center and bonds to it, expelling another group called a leaving group. Thus the incoming group replaces the leaving group in one step...
is clearly observed in the solved structure of SERCA with ADP
Adenosine diphosphate
Adenosine diphosphate, abbreviated ADP, is a nucleoside diphosphate. It is an ester of pyrophosphoric acid with the nucleoside adenosine. ADP consists of the pyrophosphate group, the pentose sugar ribose, and the nucleobase adenine....
plus AlF4-.
Differences from SERCA1a
Various subfamilies of P-type ATPases also need additional subunits for proper function. Both P-IA and P-IV pumps needs extra subunits to function. The functional unit of Na+/K+-ATPase consists of two additional subunits, beta, and gamma, involved in trafficking, folding, and regulation of these pumps. SERCA1aSERCA
SERCA, or sarco/endoplasmic reticulum Ca2+-ATPase, or SR Ca2+-ATPase, is a calcium ATPase-type P-ATPase.-Function:SERCA resides in the sarcoplasmic reticulum within muscle cells...
and other P-IIA ATPases are also regulated by phospholamban and sarcolipin in vivo. Probably other subfamilies also need additional subunits for the proper function in vivo.
Some members of the family have additional domains fused to the pump. Heavy metal pumps can have several N- and C-terminal heavy metal-binding domains
HMA domain
In molecular biology, the HMA domain is a conserved protein domain found in a number of heavy metal transport or detoxification proteins....
that have been found to be involved in regulation.
The proton pumps (IIIA) have a C-terminal regulatory domain (called the R domain), which, when unphosphorylated, inhibit pumping.
While most subfamilies have 10 transmembrane helices, there are some notable exceptions. The P-IA ATPases are predicted to have 7, and the large subfamily of heavy metal pumps (IB) is predicted to have 8 transmembrane helices. Type V appears to have a total of 12 transmembrane helices.
Mechanism
All P-type ATPases use the energy derived from ATP to drive vectorial transport. They form a high-energy aspartyl-phosphoranhydride intermediate in the reaction cycle, and they interconvert between at least two different conformations, denoted by E1 and E2. The E1-E2 notation stems from the initial studies on this family of enzymes made on the Na+,K+-ATPase, where the sodium form and the potassium form are referred to as E1 and E2, respectively, in the "Post-Albers scheme".The E1-E2 schema has been proven to work, but there exist more than two major conformational states. However, the E1-E2 notation highlights the selectivity of the enzyme
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
. In E1, the pump has high affinity for the exported substrate and low affinity for the imported substrate. In E2, it has low affinity of the exported substrate and high affinity for the imported substrate.
Four major enzyme states form the cornerstones in the reaction cycle. Several additional reaction intermediates occur interposed. These are termed E1~P, E2P, E2-P*, and E1/E2, described below.
In the case of SERCA1a
SERCA
SERCA, or sarco/endoplasmic reticulum Ca2+-ATPase, or SR Ca2+-ATPase, is a calcium ATPase-type P-ATPase.-Function:SERCA resides in the sarcoplasmic reticulum within muscle cells...
, energy from ATP
Adenosine triphosphate
Adenosine-5'-triphosphate is a multifunctional nucleoside triphosphate used in cells as a coenzyme. It is often called the "molecular unit of currency" of intracellular energy transfer. ATP transports chemical energy within cells for metabolism...
is used to transport 2 Ca2+-ions from the cytoplasmic side to the lumen
Lumen (anatomy)
A lumen in biology is the inside space of a tubular structure, such as an artery or intestine...
of the sarcoplasmatic reticulum, and to countertransport 1-3 protons into the cytoplasm
Cytoplasm
The cytoplasm is a small gel-like substance residing between the cell membrane holding all the cell's internal sub-structures , except for the nucleus. All the contents of the cells of prokaryote organisms are contained within the cytoplasm...
.
Starting in the E1/E2 state, the reaction cycle begins as the enzyme releases 1-3 protons from the cation-ligating residues, in exchange for cytoplasmic Ca2+-ions. This leads to assembly of the phosphorylation site between the ATP-bound N domain and the P domain, while the A domain directs the occlusion of the bound Ca2+. In this occluded state, the Ca2+ ions are buried in a proteinacious environment with no access to either side of the membrane.
The Ca2E1~P state becomes formed through a kinase reaction, where the P domain becomes phosphorylated, producing ADP. The cleavage of the ß,-phosphordiester bond releases the gamma-phosphate from ADP and unleashes the N domain from the P domain.
This then allows the A domain to rotate towards the phosphorylation site, making a firm association with both the P and the N domain. This movement of the A domain exerts a downward push on M3-M4 and a drag on M1-M2, forcing the pump to open at the luminal side and forming the E2P state. During this transition, the transmembrane Ca2+-binding residues are forces apart, destroying the high-affinity binding site. This is in agreement with the general model form substrate translocation (cf. 1.2), showing that energy in primary transport is not used to bind the substrate but to release it again from the buried counter ions. At the same time the N domain becomes exposed to the cytosol, ready for ATP exchange at the nucleotide-binding site.
As the Ca2+ dissociate to the luminal side, the cation binding sites are neutralised by proton binding, and this make a closure of the transmembrane segments favourable. This closure is coupled to a downwards rotation of the A domain and a movement of the P domain, which then leads to the E2-P* occluded state. Meanwhile, the N domain exchanges ADP for ATP.
The P domain is dephosphorylated by the A domain, and the cycle completes when the phosphate is released from the enzyme, stimulated by the newly bound ATP, while a cytoplasmic pathway opens to exchange the protons for two new Ca2+-ions..
See also
- H+/ K+-ATPaseHydrogen potassium ATPaseGastric hydrogen potassium ATPase is also known as H+/K+ ATPase- Function and location :The gastric hydrogen potassium ATPase or H+/K+ ATPase is the proton pump of the stomach and, as such, is the enzyme primarily responsible for the acidification of the stomach contents...
- Na+/ K+-ATPase
- H+-ATPaseProton ATPaseIn the field of enzymology, the proton-ATPase is an enzyme that catalyzes the following chemical reaction:The 3 substrates of this enzyme are ATP, H2O, and H+, whereas its 3 products are ADP, phosphate, and H+....
- Ca2+-ATPaseSERCASERCA, or sarco/endoplasmic reticulum Ca2+-ATPase, or SR Ca2+-ATPase, is a calcium ATPase-type P-ATPase.-Function:SERCA resides in the sarcoplasmic reticulum within muscle cells...

