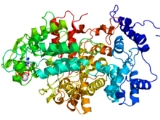
Ribonucleotide reductase
Encyclopedia
Ribonucleotide reductase (RNR, also known as ribonucleoside diphosphate reductase) is an enzyme
that catalyzes the formation of deoxyribonucleotide
s from ribonucleotide
s. Deoxyribonucleotides in turn are used in the synthesis of DNA. The reaction catalyzed by RNR is strictly conserved in all living organisms. Furthermore RNR plays a critical role in regulating the total rate of DNA synthesis so that DNA to cell mass is maintained at a constant ratio during cell division
and DNA repair
. A somewhat unusual feature of the RNR enzyme is that it catalyzes a reaction that proceeds via a free radical mechanism of action. The substrates for RNR are ADP
, GDP
, CDP
and UDP
. dTDP (deoxythymidine diphosphate) is synthesised by another enzyme (thymidylate kinase
) from dTMP (deoxythymidine monophosphate).
-dependent enzyme, ribonucleotide reductase (RNR), is essential for DNA synthesis. Class I RNR enzymes are constructed from large RNR1 and small RNR2 subunits which associate to form an active heterodimeric tetramer. Since the enzyme catalyses the de novo synthesis
of deoxyribonucleotides (dNTPs), precursors to DNA synthesis, it is essential for cell proliferation
.
In humans, the RNR1 subunit is encoded by the RRM1 gene while there are two isoforms of the RNR2 subunit, encoded by the RRM2 and RRM2B genes:
Each RNR1 monomer
consists of three domain
s:
In Pfam
, the second domain has been interpreted as two separate domains:
RNR2 contains a diferric iron center and a stable tyrosyl
radical. In E. coli
, the tyrosyl radical is located at position 122 (Y122) providing the stable radical for the Class I RNR2 subunits. In A. aegypti
, this tyrosyl radical is located at position 184 (Y184). The tyrosyl radical is deeply buried inside the protein in a hydrophobic environment, located close to the iron center that is used in the stabilization of a tyrosyl radical. The structure of two μ-oxo-linked irons is dominated by ligands that serve as iron binding sites: four carboxylates[ aspartate
(D146), glutamate
(E177, E240, and E274)] and two histidine
s (H180 and H277). Association occurs between the C-terminus of RNR2 and the C-terminus of RNR1. Enzymatic activity is dependent on association of the RNR1 and RNR2 subunits. The active site consists of the active dithiol groups from the RNR1 as well as the diferric center and the tyrosyl radical from the RNR2 subunit.
Other residues of RNR2, such as aspartate (D273), tryptophan
(W48), and tyrosine (Y356) further stabilize the active-site tyrosyl radical thus allowing electron transfer. These residues help in the transfer of the radical electron from tyrosine (Y122) of RNR2 to cysteine
(C439) of RNR1. The electron transfer begins on RNR2 tyrosine (Y122) and continues in RNR2 to tryptophan (W48), which is separated from RNR1 tyrosine (Y731) by 2.5 nanometers. Electron transfer from RNR2 to RNR1 occurs via tyrosine (Y356 to Y731) and continues on through tyrosine (Y730) to cysteine (C439) in the active site. Site-directed mutations of the RNR primary structure indicate that all residues cited above participate in the long distance transfer of the free radical to the active site.
In A. aegypti mosquitoes, RNR1 retains most of the crucial amino acid residues, including aspartate (D64) and valine (V292 or V284), that are necessary in allosteric regulation
; proline
(P210 and P610), leucine
(L453 and L473), and methionine
(M603) residues that are located in the hydrophobic active site; cysteine (C225, C436 and C451) residues that are involved in removal of a hydrogen atom and transfer of the radical electron at the active site; cysteine (C225 and C436), asparagine
(N434), and glutamate (E441) residues that bind the ribonucleotide substrate; tyrosine (Y723 and Y743) residues that dictate the radical transfer; and cysteine (C838 and C841) residues that are used in the regeneration of dithiol groups in the active site.
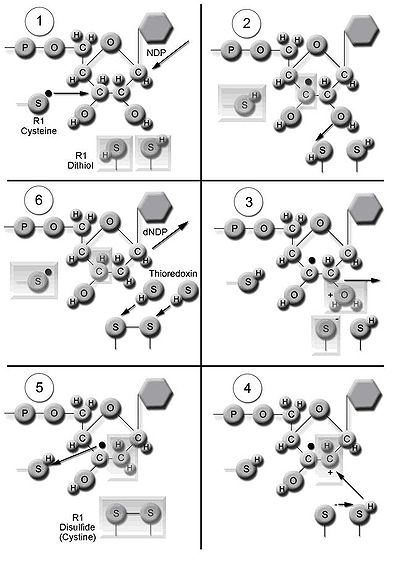 The enzyme ribonucleotide reductase (RNR) catalyzes the de novo synthesis of dNTPs. Catalysis of ribonucleoside 5’-diphosphates (NDPs) involves a reduction at the 2’-carbon of ribose 5-phosphate
The enzyme ribonucleotide reductase (RNR) catalyzes the de novo synthesis of dNTPs. Catalysis of ribonucleoside 5’-diphosphates (NDPs) involves a reduction at the 2’-carbon of ribose 5-phosphate
to form the 2’-deoxy derivative-reduced 2’-deoxyribonucleoside 5’-diphosphates (dNDPs). This reduction is initiated with the generation of a free radical. Following a single reduction, RNR requires electrons donated from the dithiol groups of the protein thioredoxin
. Regeneration of thioredoxin occurs when nicotinamide adenine dinucleotide phosphate (NADPH
) provides two hydrogen atoms that are used to reduce the disulfide
groups of thioredoxin.
Three classes of RNR have similar mechanisms for the reduction of NDPs, but differ in the domain that generates the free radical, the specific metal in the metalloprotein
structure, and the electron donors. All classes use free-radical chemistry. Class I reductases use an iron center with ferrous to ferric conversion to generate a tyrosyl free radical. Reduction of NDP substrates occurs under aerobic conditions. Class I reductases are divided into IA and IB due to differences in regulation. Class IA reductases are distributed in eukaryote
s, eubacteria
, bacteriophage
s, and virus
es. Class IB reductases are found in eubacteria. Class IB reductases can also use a radical generated with the stabilization of a binuclear manganese
center. Class II reductases generate a free radical by mechanisms involving 5’-deoxyadenosyl cobalamin (coenzyme B12) and have a simpler structure than class I and class III reductases. Reduction of NDPs or ribonucleotide 5’-triphosphates (NTPs) occurs under either aerobic
or anaerobic conditions. Class II reductases are distributed in archaebacteria
, eubacteria, and bacteriophages. Class III reductases use a glycine radical generated with the help of an S-adenosyl methionine
and an iron sulphur center. Reduction of NTPs is limited to anaerobic conditions. Class III reductases are distributed in archaebacteria, eubacteria, and bacteriophages. Organisms are not limited to having one class of enzymes. For example, E. coli have both class I and class III RNR.
. The pentose phosphate pathway produces NADPH for reducing power involved in the catalysis of NTPs to dNTPs, and to produce ribose 5-phosphate necessary for the synthesis of ribonucleotides. Carbon dioxide is always available for biosynthesis because its concentration in the blood is kept nearly constant via the bicarbonate buffer system
. An important co-factor for ribonucleotide synthesis is tetrahydrofolate, which is the major mediator for carbon transfers. Its derivative, folate (a vitamin), cannot be synthesized in mammals. Many forms of tetrahydrofolate follow pathways that are interconnected. For ribonucleotide synthesis, the N10-formyl-tetrahydrofolate molecule is necessary for the transfer of formyl groups to the purine ring. Amino groups or ammonia are donated from the catabolism of amino acids beginning with a dietary protein molecule. The free ammonia is combined with glutamate by a reaction involving adenosine 5’-triphosphate (ATP) and the activity of glutamine synthetase
, which produces a nontoxic molecule of glutamine
that can be transported in the bloodstream. Glutamine synthetase is present in nearly all organisms and is allosterically regulated by end products of glutamine metabolism. During synthesis of purine
s, amino groups are removed from glutamine for purine rings.
Purine ribonucleotides are attached to ribose 5-phosphate during assembly of intermediate inosinate
(IMP) from precursors in the purine pathway, including glutamine, glycine, N10-formyl-tetrahydrofolate, bicarbonate, aspartate and ATP. Synthesis is catalyzed by large multienzyme complexes. Purine ribonucleotides are adenosine 5’-monophosphate (AMP) and guanosine 5’-monophosphate (GMP). AMP is formed from IMP by aspartate donating an amino group (leaving as fumarate) and guanosine 5’-triphosphate (GTP) providing a phosphate. GMP is formed by the oxidation of IMP at C-2 requiring NAD+. Following oxidation, glutamine donates an amino group (leaving as glutamate) then ATP provides a phosphate.
Pyrimidine ribonucleotides are formed from an orotate molecule that is assembled from aspartate to form the pyrimidine ring. Subsequently, orotate is attached to ribose 5-phosphate to yield orotidylate. These two steps are catalyzed by a large multienzyme complex (CAD). Pyrimidine ribonucleotides are cytidine 5’-monophosphate (CMP) and uridine 5’-monophosphate (UMP). Orotidylate is decarboxylated to form UMP. UMP and two ATPs are transferred by kinases to form uridine 5’-triphosphate (UTP). Cytidine 5’-triphosphate (CTP) is formed from UTP by glutamine donating an amino group (leaving as glutamate) and ATP providing a phosphate. In some species, ammonia can donate an amino group instead of glutamine.
Generation of 2’-deoxythymidine 5’-monophosphate (dTMP) occurs by conversion of 2’-deoxyuridine 5’-monophosphate (dUMP). Thymidylate synthase catalyzes the reaction in which dTMP is formed from dUMP; to provide the carbon atom N5, N10-methylene-tetrahydrofolate is oxidized to 7, 8-dihydrofolate. Dihydrofolate reductase
(DHFR) is an essential enzyme that regenerates tetrahydrofolate at the expense of NADPH.
Ribonucleoside monophosphates (AMP, GMP, CMP, and UMP) are phosphorylated
to ribonucleoside diphosphates for their particular base by specific kinase
s. Ribonucleoside diphosphates are phosphorylated a second time to ribonucleoside triphosphates by nucleoside-diphosphate kinase
, which is not specific for their base or for their 2’-carbon of ribose 5-phosphate and its 2’-deoxy derivative. The activity of nucleoside diphosphate kinase is sequential based on which class of RNR is used. These metabolic pathways generate the ribonucleotides (ATP, GTP, CTP, and UTP) that are precursors for dNTPs. Thus, RNR reduces the corresponding NTPs to dNTPs for DNA synthesis. Cellular concentration of dNTP is much lower than required for DNA replication, and RNR is essential for adequate precursors during DNA synthesis.
After RNR reduces NDP or NTP the enzyme becomes inactive because a disulfide bond
is formed in the active site. An exchange reaction occurs that reduces the disulfide bond of RNR catalyzed by thioredoxin or glutaredoxin
. RNR gains electrons on the active-site dithiol groups necessary for its activity.
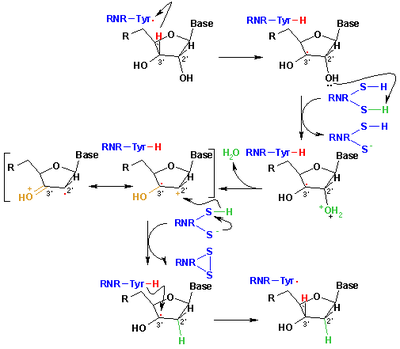 The mechanism that is currently accepted for the reduction of ribonucleotides to deoxyribonucleotides is depicted in the following scheme. The first step involves the abstraction of the 3’- H of substrate 1 by radical Cys439. Subsequently, the reaction involves the elimination of one water molecule from carbon C-2’ of the ribonucleotide, catalyzed by Cys225 and Glu441. In the third step there is a hydrogen atom transfer from Cys225 to carbon C-2’ of the 2’-ketyl radical 3, after previous proton transfer from Cys462 to Cys225. At the end of this step, a radical anionic disulfide bridge and the closed-shell ketone intermediate 4 are obtained. This intermediate has been identified during the conversion of several 2’-substituted substrate analogues, as well as with the natural substrate interacting with enzyme mutants. The next step is the oxidation of the anionic disulfide bridge, with concomitant reduction of the substrate, generating 5. The spin density shifts from the sulphur atoms to the C-3' atom of the substrate, with simultaneous proton transfer from Glu441 to carbon C-3'. The last step is the reverse of the first step and involves a hydrogen transfer from Cys439 to C-3’, regenerating the initial radical and resulting in the final product 6.
The mechanism that is currently accepted for the reduction of ribonucleotides to deoxyribonucleotides is depicted in the following scheme. The first step involves the abstraction of the 3’- H of substrate 1 by radical Cys439. Subsequently, the reaction involves the elimination of one water molecule from carbon C-2’ of the ribonucleotide, catalyzed by Cys225 and Glu441. In the third step there is a hydrogen atom transfer from Cys225 to carbon C-2’ of the 2’-ketyl radical 3, after previous proton transfer from Cys462 to Cys225. At the end of this step, a radical anionic disulfide bridge and the closed-shell ketone intermediate 4 are obtained. This intermediate has been identified during the conversion of several 2’-substituted substrate analogues, as well as with the natural substrate interacting with enzyme mutants. The next step is the oxidation of the anionic disulfide bridge, with concomitant reduction of the substrate, generating 5. The spin density shifts from the sulphur atoms to the C-3' atom of the substrate, with simultaneous proton transfer from Glu441 to carbon C-3'. The last step is the reverse of the first step and involves a hydrogen transfer from Cys439 to C-3’, regenerating the initial radical and resulting in the final product 6.
Theoretical models of some steps of these mechanisms using the full model of the R1 protein can be found at the studies performed by Cerqueira et al..
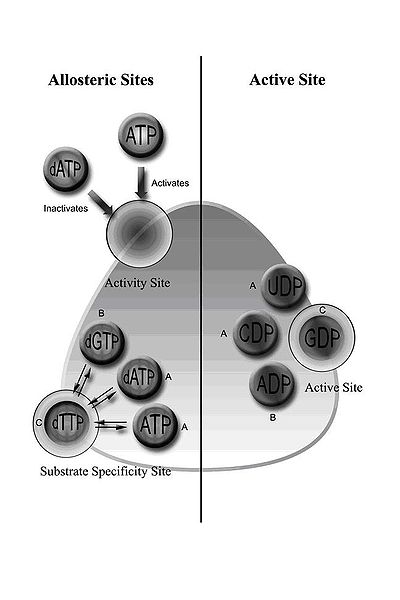 Class I RNR comprises RNR1 and RNR2 subunits, which can associate to form a heterodimeric tetramer. RNR1 contains both allosteric sites, mediating regulation of substrate specificity and activity. Depending on the allosteric configuration, one of the four ribonucleotides binds to the active site.
Class I RNR comprises RNR1 and RNR2 subunits, which can associate to form a heterodimeric tetramer. RNR1 contains both allosteric sites, mediating regulation of substrate specificity and activity. Depending on the allosteric configuration, one of the four ribonucleotides binds to the active site.
Regulation of RNR is designed to maintain balanced quantities of dNTPs. Binding of effector molecules either increases or decreases RNR activity. When ATP binds to the allosteric activity site, it activates RNR. In contrast, when dATP binds to this site, it deactivates RNR. In addition to controlling activity, the allosteric mechanism also regulates the substrate specificity and ensures the enzyme produces an equal amount of each dNTP for DNA synthesis. In all classes, binding of ATP or dATP to the allosteric site induces reduction of cytidine 5’-diphosphate (CDP) and uridine 5’-diphosphate (UDP); 2’-deoxyguanosine 5’-triphosphate (dGTP) induces reduction of adenosine 5’-diphosphate (ADP); and 2’-deoxythymidine 5’-triphosphate (dTTP) induces reduction of guanosine 5’-diphosphate (GDP) (Figure 1). Interestingly, class IB reductases are not inhibited by dATP because they lack approximately 50 N-terminal amino acids required for the allosteric activity site. Eukaryotic cells with class IA reductases have a mechanism of negative control to turn off synthesis of dNTPs as they accumulate. This mechanism protects the cell from toxic and mutagenic effects that can arise from the overproduction of dNTPs because changes in balanced dNTP pools lead to DNA damage and cell death.
Class I RNR can be inhibited by peptide
s similar to the C-terminus of RNR2. These peptides can compete with RNR2 for binding to RNR1, and as a result RNR1 does not form an enzymatically active complex with RNR2. Although the C-terminus of RNR2 proteins is different across species, RNR2 can interact with RNR1 across species. When the mouse RNR2 C-terminus was replaced with the E. coli RNR2 C-terminal (7 or 33) amino acid residues, the chimeric RNR2 subunit still binds to mouse RNR1 subunits. However, they lack enzymatic activity due probably to the elimination of residues involved in the transfer of the free radical electron from the RNR2 to the RNR1 subunit.
Small peptides can specifically inhibit the RNR2 subunits from binding with RNR1 when they share a significant similarity with the normal RNR2 C-terminus. This inhibition RNR2 binding to RNR1 has been tested successfully in herpes simplex virus (HSV) RNR. When a 7 amino acid oligomer (GAVVNDL) truncated from the C-terminus of the RNR2 subunit was used in competition assays, it prevented the normal RNR2 from forming an enzymatically active complex with RNR1. Other small peptide inhibitors similar to the RNR2 C-terminus have also been used successfully to inhibit HSV RNR enzymatic activity and thus HSV replication. In mice models of stromal
keratitis
and corneal neovascularization
(HSV
ocular disease), a small RNR2 C-terminal analog BILD 1263 has been reported to inhibit RNR and is effective in preventing these diseases. In some cases, although treatment with small C-terminal analogs may not stop disease spreading, they can still help in healing. In the acyclovir-resistant HSV (PAAr5), a small peptide inhibitor BILD 1633 has been reported to be 5 to 10 times more potent than BILD 1263 against cutaneous PAAr5 infection. A combination therapy approach (BILD 1633 and acyclovir) is more effective to heal topical lesions in mice. These data suggest that small peptide inhibitors that compete with RNR2 for binding to RNR1 are useful in preventing the spread of HSV.
Gallium
inhibits RNR2 by substituting for Fe3+ in the active site. Gallium maltolate
is an orally bioavailable form of gallium that exploits this inhibitory activity to treat cancer, infections, and other diseases.
The drugs Motexafin gadolinium
and hydroxyurea
interfere with the action of this enzyme.
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
that catalyzes the formation of deoxyribonucleotide
Deoxyribonucleotide
A deoxyribonucleotide is the monomer, or single unit, of DNA, or deoxyribonucleic acid. Each deoxyribonucleotide comprises three parts: a nitrogenous base, a deoxyribose sugar, and one phosphate group. The nitrogenous base is always bonded to the 1' carbon of the deoxyribose, which is distinguished...
s from ribonucleotide
Ribonucleotide
A ribonucleotide or ribotide is a nucleotide in which a purine or pyrimidine base is linked to a ribose molecule and exactly one phosphate group. In living organisms the most common bases for ribonucleotides are adenine , guanine , cytosine , or uracil ....
s. Deoxyribonucleotides in turn are used in the synthesis of DNA. The reaction catalyzed by RNR is strictly conserved in all living organisms. Furthermore RNR plays a critical role in regulating the total rate of DNA synthesis so that DNA to cell mass is maintained at a constant ratio during cell division
Cell division
Cell division is the process by which a parent cell divides into two or more daughter cells . Cell division is usually a small segment of a larger cell cycle. This type of cell division in eukaryotes is known as mitosis, and leaves the daughter cell capable of dividing again. The corresponding sort...
and DNA repair
DNA repair
DNA repair refers to a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as UV light and radiation can cause DNA damage, resulting in as many as 1...
. A somewhat unusual feature of the RNR enzyme is that it catalyzes a reaction that proceeds via a free radical mechanism of action. The substrates for RNR are ADP
Adenosine diphosphate
Adenosine diphosphate, abbreviated ADP, is a nucleoside diphosphate. It is an ester of pyrophosphoric acid with the nucleoside adenosine. ADP consists of the pyrophosphate group, the pentose sugar ribose, and the nucleobase adenine....
, GDP
Guanosine diphosphate
Guanosine diphosphate, abbreviated GDP, is a nucleoside diphosphate. It is an ester of pyrophosphoric acid with the nucleoside guanosine. GDP consists of the pyrophosphate group, the pentose sugar ribose, and the nucleobase guanine....
, CDP
Cytidine diphosphate
Cytidine diphosphate, abbreviated CDP, is a nucleoside diphosphate. It is an ester of pyrophosphoric acid with the nucleoside cytidine. CDP consists of the pyrophosphate group, the pentose sugar ribose, and the nucleobase cytosine....
and UDP
Uridine diphosphate
Uridine diphosphate, abbreviated UDP, is a nucleoside diphosphate. It is an ester of pyrophosphoric acid with the nucleoside uridine. UDP consists of the pyrophosphate group, the pentose sugar ribose, and the nucleobase uracil.-See also:* Nucleoside...
. dTDP (deoxythymidine diphosphate) is synthesised by another enzyme (thymidylate kinase
Thymidylate kinase
Thymidylate kinase catalyzes the phosphorylation of thymidine 5'-monophosphate to form thymidine 5'-diphosphate in the presence of ATP and magnesium:...
) from dTMP (deoxythymidine monophosphate).
Structure
The ironIron
Iron is a chemical element with the symbol Fe and atomic number 26. It is a metal in the first transition series. It is the most common element forming the planet Earth as a whole, forming much of Earth's outer and inner core. It is the fourth most common element in the Earth's crust...
-dependent enzyme, ribonucleotide reductase (RNR), is essential for DNA synthesis. Class I RNR enzymes are constructed from large RNR1 and small RNR2 subunits which associate to form an active heterodimeric tetramer. Since the enzyme catalyses the de novo synthesis
De novo synthesis
De novo synthesis refers to the synthesis of complex molecules from simple molecules such as sugars or amino acids, as opposed to their being recycled after partial degradation. For example, nucleotides are not needed in the diet as they can be constructed from small precursor molecules such as...
of deoxyribonucleotides (dNTPs), precursors to DNA synthesis, it is essential for cell proliferation
Cell growth
The term cell growth is used in the contexts of cell development and cell division . When used in the context of cell division, it refers to growth of cell populations, where one cell grows and divides to produce two "daughter cells"...
.
In humans, the RNR1 subunit is encoded by the RRM1 gene while there are two isoforms of the RNR2 subunit, encoded by the RRM2 and RRM2B genes:
Each RNR1 monomer
Monomer
A monomer is an atom or a small molecule that may bind chemically to other monomers to form a polymer; the term "monomeric protein" may also be used to describe one of the proteins making up a multiprotein complex...
consists of three domain
Protein domain
A protein domain is a part of protein sequence and structure that can evolve, function, and exist independently of the rest of the protein chain. Each domain forms a compact three-dimensional structure and often can be independently stable and folded. Many proteins consist of several structural...
s:
- one mainly helical domain comprising the 220 N-terminal residues,
- a second large ten-stranded α/β structure comprising 480 residues,
- and a third small five-stranded α/β structure comprising 70 residues.
In Pfam
Pfam
Pfam is a database of protein families that includes their annotations and multiple sequence alignments generated using hidden Markov models.- Features :For each family in Pfam one can:* Look at multiple alignments* View protein domain architectures...
, the second domain has been interpreted as two separate domains:
- a shorter all-alpha N-terminal domain,
- and a longer barrel C-terminal domain.
RNR2 contains a diferric iron center and a stable tyrosyl
Tyrosine
Tyrosine or 4-hydroxyphenylalanine, is one of the 22 amino acids that are used by cells to synthesize proteins. Its codons are UAC and UAU. It is a non-essential amino acid with a polar side group...
radical. In E. coli
Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
, the tyrosyl radical is located at position 122 (Y122) providing the stable radical for the Class I RNR2 subunits. In A. aegypti
Aedes aegypti
The yellow fever mosquito, Aedes aegypti is a mosquito that can spread the dengue fever, Chikungunya and yellow fever viruses, and other diseases. The mosquito can be recognized by white markings on legs and a marking in the form of a lyre on the thorax...
, this tyrosyl radical is located at position 184 (Y184). The tyrosyl radical is deeply buried inside the protein in a hydrophobic environment, located close to the iron center that is used in the stabilization of a tyrosyl radical. The structure of two μ-oxo-linked irons is dominated by ligands that serve as iron binding sites: four carboxylates
Aspartic acid
Aspartic acid is an α-amino acid with the chemical formula HOOCCHCH2COOH. The carboxylate anion, salt, or ester of aspartic acid is known as aspartate. The L-isomer of aspartate is one of the 20 proteinogenic amino acids, i.e., the building blocks of proteins...
(D146), glutamate
Glutamic acid
Glutamic acid is one of the 20 proteinogenic amino acids, and its codons are GAA and GAG. It is a non-essential amino acid. The carboxylate anions and salts of glutamic acid are known as glutamates...
(E177, E240, and E274)
Histidine
Histidine Histidine, an essential amino acid, has a positively charged imidazole functional group. It is one of the 22 proteinogenic amino acids. Its codons are CAU and CAC. Histidine was first isolated by German physician Albrecht Kossel in 1896. Histidine is an essential amino acid in humans...
s (H180 and H277). Association occurs between the C-terminus of RNR2 and the C-terminus of RNR1. Enzymatic activity is dependent on association of the RNR1 and RNR2 subunits. The active site consists of the active dithiol groups from the RNR1 as well as the diferric center and the tyrosyl radical from the RNR2 subunit.
Other residues of RNR2, such as aspartate (D273), tryptophan
Tryptophan
Tryptophan is one of the 20 standard amino acids, as well as an essential amino acid in the human diet. It is encoded in the standard genetic code as the codon UGG...
(W48), and tyrosine (Y356) further stabilize the active-site tyrosyl radical thus allowing electron transfer. These residues help in the transfer of the radical electron from tyrosine (Y122) of RNR2 to cysteine
Cysteine
Cysteine is an α-amino acid with the chemical formula HO2CCHCH2SH. It is a non-essential amino acid, which means that it is biosynthesized in humans. Its codons are UGU and UGC. The side chain on cysteine is thiol, which is polar and thus cysteine is usually classified as a hydrophilic amino acid...
(C439) of RNR1. The electron transfer begins on RNR2 tyrosine (Y122) and continues in RNR2 to tryptophan (W48), which is separated from RNR1 tyrosine (Y731) by 2.5 nanometers. Electron transfer from RNR2 to RNR1 occurs via tyrosine (Y356 to Y731) and continues on through tyrosine (Y730) to cysteine (C439) in the active site. Site-directed mutations of the RNR primary structure indicate that all residues cited above participate in the long distance transfer of the free radical to the active site.
In A. aegypti mosquitoes, RNR1 retains most of the crucial amino acid residues, including aspartate (D64) and valine (V292 or V284), that are necessary in allosteric regulation
Allosteric regulation
In biochemistry, allosteric regulation is the regulation of an enzyme or other protein by binding an effector molecule at the protein's allosteric site . Effectors that enhance the protein's activity are referred to as allosteric activators, whereas those that decrease the protein's activity are...
; proline
Proline
Proline is an α-amino acid, one of the twenty DNA-encoded amino acids. Its codons are CCU, CCC, CCA, and CCG. It is not an essential amino acid, which means that the human body can synthesize it. It is unique among the 20 protein-forming amino acids in that the α-amino group is secondary...
(P210 and P610), leucine
Leucine
Leucine is a branched-chain α-amino acid with the chemical formula HO2CCHCH2CH2. Leucine is classified as a hydrophobic amino acid due to its aliphatic isobutyl side chain. It is encoded by six codons and is a major component of the subunits in ferritin, astacin and other 'buffer' proteins...
(L453 and L473), and methionine
Methionine
Methionine is an α-amino acid with the chemical formula HO2CCHCH2CH2SCH3. This essential amino acid is classified as nonpolar. This amino-acid is coded by the codon AUG, also known as the initiation codon, since it indicates mRNA's coding region where translation into protein...
(M603) residues that are located in the hydrophobic active site; cysteine (C225, C436 and C451) residues that are involved in removal of a hydrogen atom and transfer of the radical electron at the active site; cysteine (C225 and C436), asparagine
Asparagine
Asparagine is one of the 20 most common natural amino acids on Earth. It has carboxamide as the side-chain's functional group. It is not an essential amino acid...
(N434), and glutamate (E441) residues that bind the ribonucleotide substrate; tyrosine (Y723 and Y743) residues that dictate the radical transfer; and cysteine (C838 and C841) residues that are used in the regeneration of dithiol groups in the active site.
Function

Ribose 5-phosphate
Ribose 5-phosphate is both a product and an intermediate of the pentose phosphate pathway. The last step of the oxidative reactions in the pentose phosphate pathway is the production of ribulose-5-phosphate. Depending on the body's state, ribulose-5-phosphate can reversibly isomerize to...
to form the 2’-deoxy derivative-reduced 2’-deoxyribonucleoside 5’-diphosphates (dNDPs). This reduction is initiated with the generation of a free radical. Following a single reduction, RNR requires electrons donated from the dithiol groups of the protein thioredoxin
Thioredoxin
Thioredoxin is a class of small redox proteins known to be present in all organisms. It plays a role in many important biological processes. In humans, it is encoded by the TXN gene. Loss-of-function mutation of either of the two human thioredoxin genes is lethal at the four-cell stage of the...
. Regeneration of thioredoxin occurs when nicotinamide adenine dinucleotide phosphate (NADPH
Nicotinamide adenine dinucleotide phosphate
Nicotinamide adenine dinucleotide phosphate, abbreviated NADP or TPN in older notation , is a coenzyme used in anabolic reactions, such as lipid and nucleic acid synthesis, which require NADPH as a reducing agent....
) provides two hydrogen atoms that are used to reduce the disulfide
Disulfide
In chemistry, a disulfide usually refers to the structural unit composed of a linked pair of sulfur atoms. Disulfide usually refer to a chemical compound that contains a disulfide bond, such as diphenyl disulfide, C6H5S-SC6H5....
groups of thioredoxin.
Three classes of RNR have similar mechanisms for the reduction of NDPs, but differ in the domain that generates the free radical, the specific metal in the metalloprotein
Metalloprotein
Metalloprotein is a generic term for a protein that contains a metal ion cofactor. Metalloproteins have many different functions in cells, such as enzymes, transport and storage proteins, and signal transduction proteins. Indeed, about one quarter to one third of all proteins require metals to...
structure, and the electron donors. All classes use free-radical chemistry. Class I reductases use an iron center with ferrous to ferric conversion to generate a tyrosyl free radical. Reduction of NDP substrates occurs under aerobic conditions. Class I reductases are divided into IA and IB due to differences in regulation. Class IA reductases are distributed in eukaryote
Eukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
s, eubacteria
Bacteria
Bacteria are a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a wide range of shapes, ranging from spheres to rods and spirals...
, bacteriophage
Bacteriophage
A bacteriophage is any one of a number of viruses that infect bacteria. They do this by injecting genetic material, which they carry enclosed in an outer protein capsid...
s, and virus
Virus
A virus is a small infectious agent that can replicate only inside the living cells of organisms. Viruses infect all types of organisms, from animals and plants to bacteria and archaea...
es. Class IB reductases are found in eubacteria. Class IB reductases can also use a radical generated with the stabilization of a binuclear manganese
Manganese
Manganese is a chemical element, designated by the symbol Mn. It has the atomic number 25. It is found as a free element in nature , and in many minerals...
center. Class II reductases generate a free radical by mechanisms involving 5’-deoxyadenosyl cobalamin (coenzyme B12) and have a simpler structure than class I and class III reductases. Reduction of NDPs or ribonucleotide 5’-triphosphates (NTPs) occurs under either aerobic
Oxygen
Oxygen is the element with atomic number 8 and represented by the symbol O. Its name derives from the Greek roots ὀξύς and -γενής , because at the time of naming, it was mistakenly thought that all acids required oxygen in their composition...
or anaerobic conditions. Class II reductases are distributed in archaebacteria
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
, eubacteria, and bacteriophages. Class III reductases use a glycine radical generated with the help of an S-adenosyl methionine
S-Adenosyl methionine
S-Adenosyl methionine is a common cosubstrate involved in methyl group transfers. SAM was first discovered in Italy by G. L. Cantoni in 1952. It is made from adenosine triphosphate and methionine by methionine adenosyltransferase . Transmethylation, transsulfuration, and aminopropylation are the...
and an iron sulphur center. Reduction of NTPs is limited to anaerobic conditions. Class III reductases are distributed in archaebacteria, eubacteria, and bacteriophages. Organisms are not limited to having one class of enzymes. For example, E. coli have both class I and class III RNR.
Metabolic pathways
Several major pathways lead to the generation of precursors for the de novo synthesis of nucleotides. These pathways involve the generation of ribose 5-phosphate, carbon dioxide, amino acids and ammonia. Ribose 5-phosphate generation begins with a molecule of glucose that is oxidized via the pentose phosphate pathwayPentose phosphate pathway
The pentose phosphate pathway is a process that generates NADPH and pentoses . There are two distinct phases in the pathway. The first is the oxidative phase, in which NADPH is generated, and the second is the non-oxidative synthesis of 5-carbon sugars...
. The pentose phosphate pathway produces NADPH for reducing power involved in the catalysis of NTPs to dNTPs, and to produce ribose 5-phosphate necessary for the synthesis of ribonucleotides. Carbon dioxide is always available for biosynthesis because its concentration in the blood is kept nearly constant via the bicarbonate buffer system
Bicarbonate buffering system
The bicarbonate buffering system is an important buffer system in the acid-base homeostasis of living things, including humans. As a buffer, it tends to maintain a relatively constant plasma pH and counteract any force that would alter it....
. An important co-factor for ribonucleotide synthesis is tetrahydrofolate, which is the major mediator for carbon transfers. Its derivative, folate (a vitamin), cannot be synthesized in mammals. Many forms of tetrahydrofolate follow pathways that are interconnected. For ribonucleotide synthesis, the N10-formyl-tetrahydrofolate molecule is necessary for the transfer of formyl groups to the purine ring. Amino groups or ammonia are donated from the catabolism of amino acids beginning with a dietary protein molecule. The free ammonia is combined with glutamate by a reaction involving adenosine 5’-triphosphate (ATP) and the activity of glutamine synthetase
Glutamine synthetase
Glutamine synthetase is an enzyme that plays an essential role in the metabolism of nitrogen by catalyzing the condensation of glutamate and ammonia to form glutamine:Glutamate + ATP + NH3 → Glutamine + ADP + phosphate...
, which produces a nontoxic molecule of glutamine
Glutamine
Glutamine is one of the 20 amino acids encoded by the standard genetic code. It is not recognized as an essential amino acid but may become conditionally essential in certain situations, including intensive athletic training or certain gastrointestinal disorders...
that can be transported in the bloodstream. Glutamine synthetase is present in nearly all organisms and is allosterically regulated by end products of glutamine metabolism. During synthesis of purine
Purine
A purine is a heterocyclic aromatic organic compound, consisting of a pyrimidine ring fused to an imidazole ring. Purines, including substituted purines and their tautomers, are the most widely distributed kind of nitrogen-containing heterocycle in nature....
s, amino groups are removed from glutamine for purine rings.
Purine ribonucleotides are attached to ribose 5-phosphate during assembly of intermediate inosinate
Inosinic acid
Inosinic acid or inosine monophosphate is a nucleotide monophosphate. Inosinic acid is important in metabolism. It is the ribonucleotide of hypoxanthine and the first nucleotide formed during the synthesis of purine. It is formed by the deamination of adenosine monophosphate, and is hydrolysed...
(IMP) from precursors in the purine pathway, including glutamine, glycine, N10-formyl-tetrahydrofolate, bicarbonate, aspartate and ATP. Synthesis is catalyzed by large multienzyme complexes. Purine ribonucleotides are adenosine 5’-monophosphate (AMP) and guanosine 5’-monophosphate (GMP). AMP is formed from IMP by aspartate donating an amino group (leaving as fumarate) and guanosine 5’-triphosphate (GTP) providing a phosphate. GMP is formed by the oxidation of IMP at C-2 requiring NAD+. Following oxidation, glutamine donates an amino group (leaving as glutamate) then ATP provides a phosphate.
Pyrimidine ribonucleotides are formed from an orotate molecule that is assembled from aspartate to form the pyrimidine ring. Subsequently, orotate is attached to ribose 5-phosphate to yield orotidylate. These two steps are catalyzed by a large multienzyme complex (CAD). Pyrimidine ribonucleotides are cytidine 5’-monophosphate (CMP) and uridine 5’-monophosphate (UMP). Orotidylate is decarboxylated to form UMP. UMP and two ATPs are transferred by kinases to form uridine 5’-triphosphate (UTP). Cytidine 5’-triphosphate (CTP) is formed from UTP by glutamine donating an amino group (leaving as glutamate) and ATP providing a phosphate. In some species, ammonia can donate an amino group instead of glutamine.
Generation of 2’-deoxythymidine 5’-monophosphate (dTMP) occurs by conversion of 2’-deoxyuridine 5’-monophosphate (dUMP). Thymidylate synthase catalyzes the reaction in which dTMP is formed from dUMP; to provide the carbon atom N5, N10-methylene-tetrahydrofolate is oxidized to 7, 8-dihydrofolate. Dihydrofolate reductase
Dihydrofolate reductase
- Function :Dihydrofolate reductase converts dihydrofolate into tetrahydrofolate, a methyl group shuttle required for the de novo synthesis of purines, thymidylic acid, and certain amino acids...
(DHFR) is an essential enzyme that regenerates tetrahydrofolate at the expense of NADPH.
Ribonucleoside monophosphates (AMP, GMP, CMP, and UMP) are phosphorylated
Phosphorylation
Phosphorylation is the addition of a phosphate group to a protein or other organic molecule. Phosphorylation activates or deactivates many protein enzymes....
to ribonucleoside diphosphates for their particular base by specific kinase
Kinase
In chemistry and biochemistry, a kinase is a type of enzyme that transfers phosphate groups from high-energy donor molecules, such as ATP, to specific substrates, a process referred to as phosphorylation. Kinases are part of the larger family of phosphotransferases...
s. Ribonucleoside diphosphates are phosphorylated a second time to ribonucleoside triphosphates by nucleoside-diphosphate kinase
Nucleoside-diphosphate kinase
Nucleoside-diphosphate kinases are enzymes that catalyze the exchange of phosphate groups between different nucleoside diphosphates...
, which is not specific for their base or for their 2’-carbon of ribose 5-phosphate and its 2’-deoxy derivative. The activity of nucleoside diphosphate kinase is sequential based on which class of RNR is used. These metabolic pathways generate the ribonucleotides (ATP, GTP, CTP, and UTP) that are precursors for dNTPs. Thus, RNR reduces the corresponding NTPs to dNTPs for DNA synthesis. Cellular concentration of dNTP is much lower than required for DNA replication, and RNR is essential for adequate precursors during DNA synthesis.
After RNR reduces NDP or NTP the enzyme becomes inactive because a disulfide bond
Disulfide bond
In chemistry, a disulfide bond is a covalent bond, usually derived by the coupling of two thiol groups. The linkage is also called an SS-bond or disulfide bridge. The overall connectivity is therefore R-S-S-R. The terminology is widely used in biochemistry...
is formed in the active site. An exchange reaction occurs that reduces the disulfide bond of RNR catalyzed by thioredoxin or glutaredoxin
Glutaredoxin
Glutaredoxins are small redox enzymes of approximately one hundred amino-acid residues that use glutathione as a cofactor. Glutaredoxins are oxidized by substrates, and reduced non-enzymatically by glutathione. In contrast to thioredoxins, which are reduced by thioredoxin reductase, no...
. RNR gains electrons on the active-site dithiol groups necessary for its activity.
Catalytic Reduction Mechanism

Theoretical models of some steps of these mechanisms using the full model of the R1 protein can be found at the studies performed by Cerqueira et al..
Regulation

Regulation of RNR is designed to maintain balanced quantities of dNTPs. Binding of effector molecules either increases or decreases RNR activity. When ATP binds to the allosteric activity site, it activates RNR. In contrast, when dATP binds to this site, it deactivates RNR. In addition to controlling activity, the allosteric mechanism also regulates the substrate specificity and ensures the enzyme produces an equal amount of each dNTP for DNA synthesis. In all classes, binding of ATP or dATP to the allosteric site induces reduction of cytidine 5’-diphosphate (CDP) and uridine 5’-diphosphate (UDP); 2’-deoxyguanosine 5’-triphosphate (dGTP) induces reduction of adenosine 5’-diphosphate (ADP); and 2’-deoxythymidine 5’-triphosphate (dTTP) induces reduction of guanosine 5’-diphosphate (GDP) (Figure 1). Interestingly, class IB reductases are not inhibited by dATP because they lack approximately 50 N-terminal amino acids required for the allosteric activity site. Eukaryotic cells with class IA reductases have a mechanism of negative control to turn off synthesis of dNTPs as they accumulate. This mechanism protects the cell from toxic and mutagenic effects that can arise from the overproduction of dNTPs because changes in balanced dNTP pools lead to DNA damage and cell death.
RNR1 and RNR2 inhibitors
Generally Class I RNR inhibitors can be divided in three main groups: translation inhibitors, which block the synthesis of the enzyme; dimerization inhibitors that prevent the association of the two RNR subunits (R1 and R2); and catalytic inhibitors that inactivate the subunit R1 and/or subunit R2.Class I RNR can be inhibited by peptide
Peptide
Peptides are short polymers of amino acid monomers linked by peptide bonds. They are distinguished from proteins on the basis of size, typically containing less than 50 monomer units. The shortest peptides are dipeptides, consisting of two amino acids joined by a single peptide bond...
s similar to the C-terminus of RNR2. These peptides can compete with RNR2 for binding to RNR1, and as a result RNR1 does not form an enzymatically active complex with RNR2. Although the C-terminus of RNR2 proteins is different across species, RNR2 can interact with RNR1 across species. When the mouse RNR2 C-terminus was replaced with the E. coli RNR2 C-terminal (7 or 33) amino acid residues, the chimeric RNR2 subunit still binds to mouse RNR1 subunits. However, they lack enzymatic activity due probably to the elimination of residues involved in the transfer of the free radical electron from the RNR2 to the RNR1 subunit.
Small peptides can specifically inhibit the RNR2 subunits from binding with RNR1 when they share a significant similarity with the normal RNR2 C-terminus. This inhibition RNR2 binding to RNR1 has been tested successfully in herpes simplex virus (HSV) RNR. When a 7 amino acid oligomer (GAVVNDL) truncated from the C-terminus of the RNR2 subunit was used in competition assays, it prevented the normal RNR2 from forming an enzymatically active complex with RNR1. Other small peptide inhibitors similar to the RNR2 C-terminus have also been used successfully to inhibit HSV RNR enzymatic activity and thus HSV replication. In mice models of stromal
Stroma (animal tissue)
In animal tissue, stroma refers to the connective, supportive framework of a biological cell, tissue, or organ.The stroma in animal tissue is contrasted with the parenchyma.Examples include:* Stroma of iris...
keratitis
Keratitis
Keratitis is a condition in which the eye's cornea, the front part of the eye, becomes inflamed. The condition is often marked by moderate to intense pain and usually involves impaired eyesight.-Types:...
and corneal neovascularization
Corneal neovascularization
Corneal neovascularization is the excessive ingrowth of blood vessels from the limbal vascular plexus into the cornea, caused by a low reception of oxygen, which is generally not received from the bloodstream, but through the air. One of the most common causes is contact lens wear, and to a...
(HSV
Herpes simplex
Herpes simplex is a viral disease caused by both Herpes simplex virus type 1 and type 2 . Infection with the herpes virus is categorized into one of several distinct disorders based on the site of infection. Oral herpes, the visible symptoms of which are colloquially called cold sores or fever...
ocular disease), a small RNR2 C-terminal analog BILD 1263 has been reported to inhibit RNR and is effective in preventing these diseases. In some cases, although treatment with small C-terminal analogs may not stop disease spreading, they can still help in healing. In the acyclovir-resistant HSV (PAAr5), a small peptide inhibitor BILD 1633 has been reported to be 5 to 10 times more potent than BILD 1263 against cutaneous PAAr5 infection. A combination therapy approach (BILD 1633 and acyclovir) is more effective to heal topical lesions in mice. These data suggest that small peptide inhibitors that compete with RNR2 for binding to RNR1 are useful in preventing the spread of HSV.
Gallium
Gallium
Gallium is a chemical element that has the symbol Ga and atomic number 31. Elemental gallium does not occur in nature, but as the gallium salt in trace amounts in bauxite and zinc ores. A soft silvery metallic poor metal, elemental gallium is a brittle solid at low temperatures. As it liquefies...
inhibits RNR2 by substituting for Fe3+ in the active site. Gallium maltolate
Gallium maltolate
Gallium maltolate is a coordination complex consisting of a trivalent gallium cation coordinated to three maltolate ligands. The compound is undergoing clinical and preclinical testing as a potential therapeutic agent for cancer, infectious disease, and inflammatory disease...
is an orally bioavailable form of gallium that exploits this inhibitory activity to treat cancer, infections, and other diseases.
The drugs Motexafin gadolinium
Motexafin gadolinium
Motexafin gadolinium is an inhibitor of thioredoxin reductase and ribonucleotide reductase. It has been proposed as a possible chemotherapeutic agent in the treatment of brain cancer.-History:...
and hydroxyurea
Hydroxyurea
Hydroxycarbamide or hydroxyurea is an antineoplastic drug, first synthesized in 1869, used in myeloproliferative disorders, specifically polycythemia vera and essential thrombocythemia...
interfere with the action of this enzyme.

