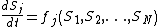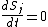Gene regulatory network
Encyclopedia
Gene
A gene is a molecular unit of heredity of a living organism. It is a name given to some stretches of DNA and RNA that code for a type of protein or for an RNA chain that has a function in the organism. Living beings depend on genes, as they specify all proteins and functional RNA chains...
regulatory network or genetic regulatory network (GRN) is a collection of DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
segments in a cell which
interact with each other indirectly (through their RNA and protein expression products) and with other substances in the cell, thereby governing the rates at which genes in the network are transcribed into mRNA.
In general, each mRNA molecule goes on to make a specific protein (or set of proteins). In some cases this protein will be structural, and will accumulate at the cell membrane or within the cell to give it particular structural properties. In other cases the protein will be an enzyme
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
, i.e., a micro-machine that catalyses a certain reaction, such as the breakdown of a food source or toxin. Some proteins though serve only to activate other genes, and these are the transcription factors that are the main players in regulatory networks or cascades. By binding to the promoter region at the start of other genes they turn them on, initiating the production of another protein, and so on. Some transcription factors are inhibitory.
In single-celled organisms, regulatory networks respond to the external environment, optimising the cell at a given time for survival in this environment. Thus a yeast cell, finding itself in a sugar solution, will turn on genes to make enzymes that process the sugar to alcohol. This process, which we associate with wine-making, is how the yeast cell makes its living, gaining energy to multiply, which under normal circumstances would enhance its survival prospects.
In multicellular animals the same principle has been put in the service of gene cascades that control body-shape. Each time a cell divides, two cells result which, although they contain the same genome in full, can differ in which genes are turned on and making proteins. Sometimes a 'self-sustaining feedback loop' ensures that a cell maintains its identity and passes it on. Less understood is the mechanism of epigenetics
Epigenetics
In biology, and specifically genetics, epigenetics is the study of heritable changes in gene expression or cellular phenotype caused by mechanisms other than changes in the underlying DNA sequence – hence the name epi- -genetics...
by which chromatin
Chromatin
Chromatin is the combination of DNA and proteins that make up the contents of the nucleus of a cell. The primary functions of chromatin are; to package DNA into a smaller volume to fit in the cell, to strengthen the DNA to allow mitosis and meiosis and prevent DNA damage, and to control gene...
modification may provide cellular memory by blocking or allowing transcription. A major feature of multicellular animals is the use of morphogen
Morphogen
A morphogen is a substance governing the pattern of tissue development, and the positions of the various specialized cell types within a tissue...
gradients, which in effect provide a positioning system that tells a cell where in the body it is, and hence what sort of cell to become. A gene that is turned on in one cell may make a product that leaves the cell and diffuses through adjacent cells, entering them and turning on genes only when it is present above a certain threshold level. These cells are thus induced into a new fate, and may even generate other morphogens that signal back to the original cell. Over longer distances morphogens may use the active process of signal transduction
Signal transduction
Signal transduction occurs when an extracellular signaling molecule activates a cell surface receptor. In turn, this receptor alters intracellular molecules creating a response...
. Such signalling controls embryogenesis
Embryogenesis
Embryogenesis is the process by which the embryo is formed and develops, until it develops into a fetus.Embryogenesis starts with the fertilization of the ovum by sperm. The fertilized ovum is referred to as a zygote...
, the building of a body plan
Body plan
A body plan is the blueprint for the way the body of an organism is laid out. An organism's symmetry, its number of body segments and number of limbs are all aspects of its body plan...
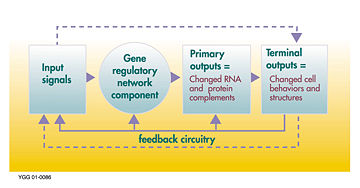
from scratch through a series of sequential steps. They also control maintain adult bodies through feedback
Feedback
Feedback describes the situation when output from an event or phenomenon in the past will influence an occurrence or occurrences of the same Feedback describes the situation when output from (or information about the result of) an event or phenomenon in the past will influence an occurrence or...
processes, and the loss of such feedback because of a mutation can be responsible for the cell proliferation that is seen in cancer
Cancer
Cancer , known medically as a malignant neoplasm, is a large group of different diseases, all involving unregulated cell growth. In cancer, cells divide and grow uncontrollably, forming malignant tumors, and invade nearby parts of the body. The cancer may also spread to more distant parts of the...
. In parallel with this process of building structure, the gene cascade turns on genes that make structural proteins that give each cell the physical properties it needs. It has been suggested that, because biological molecular interactions are intrinsically stochastic, gene networks are the result of cellular processes and not their cause (i.e. Cellular Darwinism). However, recent experimental evidence has favored the attractor view of cell fates.
Overview
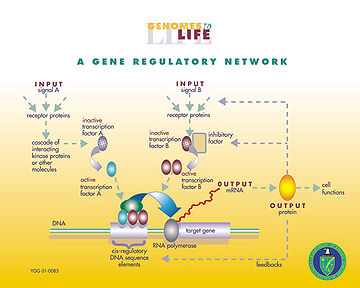
At one level, biological cells can be thought of as "partially-mixed bags" of biological chemicals – in the discussion of gene regulatory networks, these chemicals are mostly the mRNAs and proteins that arise from gene expression. These mRNA and proteins interact with each other with various degrees of specificity. Some diffuse around the cell. Others are bound to cell membranes, interacting with molecules in the environment. Still others pass through cell membranes and mediate long range signals to other cells in a multi-cellular organism. These molecules and their interactions comprise a gene regulatory network. A typical gene regulatory network looks something like this:The nodes of this network are proteins, their corresponding mRNAs, and protein/protein complexes. Nodes that are depicted as lying along vertical lines are associated with the cell/environment interfaces, while the others are free-floating and diffusible. Implied are genes, the DNA sequences which are transcribed into the mRNAs that translate into proteins. Edges between nodes represent individual molecular reactions, the protein/protein and protein/mRNA interactions through which the products of one gene affect those of another, though the lack of experimentally obtained information often implies that some reactions are not modeled at such a fine level of detail. These interactions can be inductive (the arrowheads), with an increase in the concentration of one leading to an increase in the other, or inhibitory (the filled circles), with an increase in one leading to a decrease in the other. A series of edges indicates a chain of such dependences, with cycles corresponding to feedback loops. The network structure is an abstraction of the system's chemical dynamics, describing the manifold ways in which one substance affects all the others to which it is connected. In practice, such GRNs are inferred from the biological literature on a given system and represent a distillation of the collective knowledge about a set of related biochemical reactions.
Genes can be viewed as nodes in the network, with input being proteins such as transcription factor
Transcription factor
In molecular biology and genetics, a transcription factor is a protein that binds to specific DNA sequences, thereby controlling the flow of genetic information from DNA to mRNA...
s, and outputs being the level of gene expression
Gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA , transfer RNA or small nuclear RNA genes, the product is a functional RNA...
. The node itself can also be viewed as a function which can be obtained by combining basic functions upon the inputs (in the Boolean network described below these are Boolean functions, typically AND, OR, and NOT). These functions have been interpreted as performing a kind of information processing
Information processing
Information processing is the change of information in any manner detectable by an observer. As such, it is a process which describes everything which happens in the universe, from the falling of a rock to the printing of a text file from a digital computer system...
within the cell, which determines cellular behavior. The basic drivers within cells are concentrations of some proteins, which determine both spatial (location within the cell or tissue) and temporal (cell cycle or developmental stage) coordinates of the cell, as a kind of "cellular memory". The gene networks are only beginning to be understood, and it is a next step for biology to attempt to deduce the functions for each gene "node", to help understand the behavior of the system in increasing levels of complexity, from gene to signaling pathway, cell or tissue level (see systems biology
Systems biology
Systems biology is a term used to describe a number of trends in bioscience research, and a movement which draws on those trends. Proponents describe systems biology as a biology-based inter-disciplinary study field that focuses on complex interactions in biological systems, claiming that it uses...
).
Mathematical model
Mathematical model
A mathematical model is a description of a system using mathematical concepts and language. The process of developing a mathematical model is termed mathematical modeling. Mathematical models are used not only in the natural sciences and engineering disciplines A mathematical model is a...
s of GRNs have been developed to capture the behavior of the system being modeled, and in some cases generate predictions corresponding with experimental observations. In some other cases, models have proven to make accurate novel predictions, which can be tested experimentally, thus suggesting new approaches to explore in an experiment that sometimes wouldn't be considered in the design of the protocol of an experimental laboratory. The most common modeling technique involves the use of coupled ordinary differential equation
Differential equation
A differential equation is a mathematical equation for an unknown function of one or several variables that relates the values of the function itself and its derivatives of various orders...
s (ODEs). Several other promising modeling techniques have been used, including Boolean network
Boolean network
A Boolean network consists of a set of Boolean variables whose state is determined by other variables in the network. They are a particular case of discrete dynamical networks, where time and states are discrete, i.e. they have a bijection onto an integer series...
s, Petri net
Petri net
A Petri net is one of several mathematical modeling languages for the description of distributed systems. A Petri net is a directed bipartite graph, in which the nodes represent transitions and places...
s, Bayesian network
Bayesian network
A Bayesian network, Bayes network, belief network or directed acyclic graphical model is a probabilistic graphical model that represents a set of random variables and their conditional dependencies via a directed acyclic graph . For example, a Bayesian network could represent the probabilistic...
s, graphical Gaussian models, Stochastic
Stochastic
Stochastic refers to systems whose behaviour is intrinsically non-deterministic. A stochastic process is one whose behavior is non-deterministic, in that a system's subsequent state is determined both by the process's predictable actions and by a random element. However, according to M. Kac and E...
, and Process Calculi
Process calculus
In computer science, the process calculi are a diverse family of related approaches for formally modelling concurrent systems. Process calculi provide a tool for the high-level description of interactions, communications, and synchronizations between a collection of independent agents or processes...
. Conversely, techniques have been proposed for generating models of GRNs that best explain a set of time series
Time series
In statistics, signal processing, econometrics and mathematical finance, a time series is a sequence of data points, measured typically at successive times spaced at uniform time intervals. Examples of time series are the daily closing value of the Dow Jones index or the annual flow volume of the...
observations.
Coupled ODEs
It is common to model such a network with a set of coupled ordinary differential equationOrdinary differential equation
In mathematics, an ordinary differential equation is a relation that contains functions of only one independent variable, and one or more of their derivatives with respect to that variable....
s (ODEs) or stochastic ODE
Stochastic differential equation
A stochastic differential equation is a differential equation in which one or more of the terms is a stochastic process, thus resulting in a solution which is itself a stochastic process....
s, describing the reaction kinetics of the constituent parts. Suppose that our regulatory network has
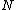 nodes, and let
nodes, and let 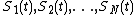 represent the concentrations of the
represent the concentrations of the 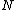 corresponding substances at time
corresponding substances at time  . Then the temporal evolution of the system can be described approximately by
. Then the temporal evolution of the system can be described approximately bywhere the functions
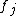 express the dependence of
express the dependence of 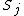 on the concentrations of other substances present in the cell. The functions
on the concentrations of other substances present in the cell. The functions 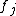 are ultimately derived from basic principles of chemical kinetics
are ultimately derived from basic principles of chemical kineticsRate equation
The rate law or rate equation for a chemical reaction is an equation that links the reaction rate with concentrations or pressures of reactants and constant parameters . To determine the rate equation for a particular system one combines the reaction rate with a mass balance for the system...
or simple expressions derived from these e.g. Michaelis-Menten enzymatic kinetics. Hence, the functional forms of the
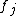 are usually chosen as low-order polynomials or Hill functions that serve as an ansatz
are usually chosen as low-order polynomials or Hill functions that serve as an ansatzAnsatz
Ansatz is a German noun with several meanings in the English language.It is widely encountered in physics and mathematics literature.Since ansatz is a noun, in German texts the initial a of this word is always capitalised.-Definition:...
for the real molecular dynamics. Such models are then studied using the mathematics of nonlinear dynamics
Dynamical system
A dynamical system is a concept in mathematics where a fixed rule describes the time dependence of a point in a geometrical space. Examples include the mathematical models that describe the swinging of a clock pendulum, the flow of water in a pipe, and the number of fish each springtime in a...
. System-specific information, like reaction rate
Reaction rate
The reaction rate or speed of reaction for a reactant or product in a particular reaction is intuitively defined as how fast or slow a reaction takes place...
constants and sensitivities, are encoded as constant parameters.
By solving for the fixed point
Fixed point (mathematics)
In mathematics, a fixed point of a function is a point that is mapped to itself by the function. A set of fixed points is sometimes called a fixed set...
of the system:
for all
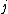 , one obtains (possibly several) concentration profiles of proteins and mRNAs that are theoretically sustainable (though not necessarily stable). Steady state
, one obtains (possibly several) concentration profiles of proteins and mRNAs that are theoretically sustainable (though not necessarily stable). Steady stateSteady state
A system in a steady state has numerous properties that are unchanging in time. This implies that for any property p of the system, the partial derivative with respect to time is zero:...
s of kinetic equations thus correspond to potential cell types, and oscillatory
Oscillation
Oscillation is the repetitive variation, typically in time, of some measure about a central value or between two or more different states. Familiar examples include a swinging pendulum and AC power. The term vibration is sometimes used more narrowly to mean a mechanical oscillation but sometimes...
solutions to the above equation to naturally cyclic cell types. Mathematical stability of these attractor
Attractor
An attractor is a set towards which a dynamical system evolves over time. That is, points that get close enough to the attractor remain close even if slightly disturbed...
s can usually be characterized by the sign of higher derivatives at critical points, and then correspond to biochemical stability
Steady state (biochemistry)
In ionic steady state, cells maintain different internal and external concentrations of various ionic species. Cell membranes are permeable to sodium and various other ions, so in order to maintain a constant ionic concentration the cell must expend energy to actively transport these ions against...
of the concentration profile. Critical point
Critical point (mathematics)
In calculus, a critical point of a function of a real variable is any value in the domain where either the function is not differentiable or its derivative is 0. The value of the function at a critical point is a critical value of the function...
s and bifurcation
Bifurcation theory
Bifurcation theory is the mathematical study of changes in the qualitative or topological structure of a given family, such as the integral curves of a family of vector fields, and the solutions of a family of differential equations...
s in the equations correspond to critical cell states in which small state or parameter perturbations could switch the system between one of several stable differentiation fates. Trajectories correspond to the unfolding of biological pathways and transients of the equations to short-term biological events. For a more mathematical discussion, see the articles on nonlinearity
Nonlinearity
In mathematics, a nonlinear system is one that does not satisfy the superposition principle, or one whose output is not directly proportional to its input; a linear system fulfills these conditions. In other words, a nonlinear system is any problem where the variable to be solved for cannot be...
, dynamical systems, bifurcation theory
Bifurcation theory
Bifurcation theory is the mathematical study of changes in the qualitative or topological structure of a given family, such as the integral curves of a family of vector fields, and the solutions of a family of differential equations...
, and chaos theory
Chaos theory
Chaos theory is a field of study in mathematics, with applications in several disciplines including physics, economics, biology, and philosophy. Chaos theory studies the behavior of dynamical systems that are highly sensitive to initial conditions, an effect which is popularly referred to as the...
.
Boolean network
The following example illustrates how a Boolean networkBoolean network
A Boolean network consists of a set of Boolean variables whose state is determined by other variables in the network. They are a particular case of discrete dynamical networks, where time and states are discrete, i.e. they have a bijection onto an integer series...
can model a GRN together with its gene products (the outputs) and the substances from the environment that affect it (the inputs). Stuart Kauffman
Stuart Kauffman
Stuart Alan Kauffman is an American theoretical biologist and complex systems researcher concerning the origin of life on Earth...
was amongst the first biologists to use the metaphor of Boolean networks to model genetic regulatory networks.
- Each gene, each input, and each output is represented by a node in a directed graphDirected graphA directed graph or digraph is a pair G= of:* a set V, whose elements are called vertices or nodes,...
in which there is an arrow from one node to another if and only if there is a causal link between the two nodes. - Each node in the graph can be in one of two states: on or off.
- For a gene, "on" corresponds to the gene being expressed; for inputs and outputs, "on" corresponds to the substance being present.
- Time is viewed as proceeding in discrete steps. At each step, the new state of a node is a Boolean function of the prior states of the nodes with arrows pointing towards it.
The validity of the model can be tested by comparing simulation results with time series observations.
Continuous networks
Continuous network models of GRNs are an extension of the boolean networks described above. Nodes still represent genes and connections between them regulatory influences on gene expression. Genes in biological systems display a continuous range of activity levels and it has been argued that using a continuous representation captures several properties of gene regulatory networks not present in the Boolean model. Formally most of these approaches are similar to an artificial neural networkArtificial neural network
An artificial neural network , usually called neural network , is a mathematical model or computational model that is inspired by the structure and/or functional aspects of biological neural networks. A neural network consists of an interconnected group of artificial neurons, and it processes...
, as inputs to a node are summed up and the result serves as input to a sigmoid function, e.g., but proteins do often control gene expression in a synergistic, i.e. non-linear, way. However there is now a continuous network model that allows grouping of inputs to a node thus realizing another level of regulation. This model is formally closer to a higher order recurrent neural network
Recurrent neural network
A recurrent neural network is a class of neural network where connections between units form a directed cycle. This creates an internal state of the network which allows it to exhibit dynamic temporal behavior. Unlike feedforward neural networks, RNNs can use their internal memory to process...
. The same model has also been used to mimic the evolution of cellular differentiation
Cellular differentiation
In developmental biology, cellular differentiation is the process by which a less specialized cell becomes a more specialized cell type. Differentiation occurs numerous times during the development of a multicellular organism as the organism changes from a simple zygote to a complex system of...
and even multicellular morphogenesis
Morphogenesis
Morphogenesis , is the biological process that causes an organism to develop its shape...
.
Stochastic gene networks
Recent experimental resultshave demonstrated that gene expression is a stochastic process. Thus, many authors are now using the stochastic formalism, after the work by. Works on single gene expression and small synthetic genetic networks, such as the genetic toggle switch of Tim Gardner and Jim Collins
James Collins (Boston University)
James J. Collins is an American bioengineer, Professor of Biomedical Engineering at Boston University, and a Howard Hughes Medical Institute Investigator...
, provided additional experimental data on the phenotypic variability and the stochastic nature of gene expression. The first versions of stochastic models of gene expression involved only instantaneous reactions and were driven by the Gillespie algorithm
Gillespie algorithm
In probability theory, the Gillespie algorithm generates a statistically correct trajectory of a stochastic equation. It was created by Joseph L...
.
Since some processes, such as gene transcription, involve many reactions and could not be correctly modeled as an instantaneous reaction in a single step, it was proposed to model these reactions as single step multiple delayed reactions in order to account for the time it takes for the entire process to be complete.
From here, a set of reactions were proposed that allow generating GRNs. These are then simulated using a modified version of the Gillespie algorithm, that can simulate multiple time delayed reactions (chemical reactions where each of the products is provided a time delay that determines when will it be released in the system as a "finished product").
For example, basic transcription of a gene can be represented by the following single-step reaction (RNAP is the RNA polymerase, RBS is the RNA ribosome binding site, and Pro i is the promoter region of gene i):
-

A recent work proposed a simulator (SGNSim, Stochastic Gene Networks Simulator), that can model GRNs where transcription and translation are modeled as multiple time delayed events and its dynamics is driven by a stochastic simulation algorithm (SSA) able to deal with multiple time delayed events.
The time delays can be drawn from several distributions and the reaction rates from complex
functions or from physical parameters. SGNSim can generate ensembles of GRNs within a set of user-defined parameters, such as topology. It can also be used to model specific GRNs and systems of chemical reactions. Genetic perturbations such as gene deletions, gene over-expression, insertions, frame shift mutations can also be modeled as well.
The GRN is created from a graph with the desired topology, imposing in-degree and out-degree distributions. Gene promoter activities are affected by other genes expression products that act as inputs, in the form of monomers or combined into multimers and set as direct or indirect. Next, each direct input is assigned to an operator site and different transcription factors can be allowed, or not, to compete for the same operator site, while indirect inputs are given a target. Finally, a function is assigned to each gene, defining the gene's response to a combination of transcription factors (promoter state). The transfer functions (that is, how genes respond to a combination of inputs) can be assigned to each combination of promoter states as desired.
In other recent work, multiscale models of gene regulatory networks have been developed that focus on synthetic biology applications. Simulations have been used that model all biomolecular interactions in transcription, translation, regulation, and induction of gene regulatory networks, guiding the design of synthetic systems.
Prediction
Other work has focused on predicting the gene expression levels in a gene regulatory network. The approaches used to model gene regulatory networks have been constrained to be interpretable and, as a result, are generally simplified versions of the network. For example, Boolean networks have been used due to their simplicity and ability to handle noisy data but lose data information by having a binary representation of the genes. Also, artificial neural networks omit using a hidden layer so that they can be interpreted, losing the ability to model higher order correlations in the data. Using a model that is not constrained to be interpretable, a more accurate model can be produced. Being able to predict gene expressions more accurately provides a way to explore how drugs affect a system of genes as well as for finding which genes are interrelated in a process. This has been encouraged by the DREAM competition which promotes a competition for the best prediction algorithms. Some other recent work has used artificial neural networks with a hidden layer.
Network connectivity
Empirical data indicate that biological gene networks are sparsely connected, and that the average number of upstream-regulators per gene is less than two. Theoretical results show that selection for robust gene networks will favor minimally complex, more sparsely connected, networks. These results suggest that a sparse, minimally connected, genetic architecture may be a fundamental design constraint shaping the evolution of gene network complexity.
See also
- Body planBody planA body plan is the blueprint for the way the body of an organism is laid out. An organism's symmetry, its number of body segments and number of limbs are all aspects of its body plan...
- Cis-regulatory moduleCis-regulatory moduleCis-regulatory module is a stretch of DNA, usually 100-1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes. One cis-regulatory element can regulate several genes, and conversely, one gene can have several cis-regulatory modules...
- GenenetworkGenenetworkGeneNetwork is a database and open source bioinformatics software resource for systems genetics. This resource is used to study gene regulatory networks that link DNA sequence variants to corresponding differences in gene and protein expression and to differences in traits such as health and...
(database) - MorphogenMorphogenA morphogen is a substance governing the pattern of tissue development, and the positions of the various specialized cell types within a tissue...
- OperonOperonIn genetics, an operon is a functioning unit of genomic DNA containing a cluster of genes under the control of a single regulatory signal or promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo trans-splicing to create...
- SynexpressionSynexpressionSynexpression is a type of non-random eukaryotic gene organization. Genes in a synexpression group may not be physically linked, but they are involved in the same process and they are coordinately expressed. It is expected that genes that function in the same process be regulated coordinately...
- Systems biologySystems biologySystems biology is a term used to describe a number of trends in bioscience research, and a movement which draws on those trends. Proponents describe systems biology as a biology-based inter-disciplinary study field that focuses on complex interactions in biological systems, claiming that it uses...
External links
- Gene Regulatory Networks — Short introduction
- Open source web service for GRN analysis
- BIB: Yeast Biological Interaction Browser
- Graphical Gaussian models for genome data — Inference of gene association networks with GGMs
- A bibliography on learning causal networks of gene interactions - regularly updated, contains hundreds of links to papers from bioinformatics, statistics, machine learning.
- http://mips.gsf.de/proj/biorel/ BIOREL is a web-based resource for quantitative estimation of the gene network bias in relation to available database information about gene activity/function/properties/associations/interactio.
- Evolving Biological Clocks using Genetic Regulatory Networks - Information page with model source code and Java applet.
- Engineered Gene Networks
- Tutorial: Genetic Algorithms and their Application to the Artificial Evolution of Genetic Regulatory Networks
- BEN: a web-based resource for exploring the connections between genes, diseases, and other biomedical entities
- Body plan