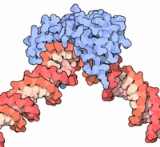
Transcription factor
Encyclopedia
In molecular biology
and genetics
, a transcription factor (sometimes called a sequence-specific DNA
-binding factor) is a protein
that binds to specific DNA sequence
s, thereby controlling the flow (or transcription
) of genetic information from DNA to mRNA
. Transcription factors perform this function alone or with other proteins in a complex, by promoting (as an activator
), or blocking (as a repressor
) the recruitment of RNA polymerase
(the enzyme that performs the transcription
of genetic information from DNA to RNA) to specific genes.
A defining feature of transcription factors is that they contain one or more DNA-binding domain
s (DBDs), which attach to specific sequences of DNA adjacent to the genes that they regulate. Additional proteins such as coactivator
s, chromatin remodeler
s, histone acetylases
, deacetylases
, kinase
s, and methylase
s, while also playing crucial roles in gene regulation
, lack DNA-binding domains, and, therefore, are not classified as transcription factors.
There are approximately 2600 proteins in the human genome
that contain DNA-binding domains, and most of these are presumed to function as transcription factors. Therefore, approximately 10% of genes in the genome code for transcription factors, which makes this family the single largest family of human proteins. Furthermore, genes are often flanked by several binding sites for distinct transcription factors, and efficient expression of each of these genes requires the cooperative action of several different transcription factors (see, for example, hepatocyte nuclear factors). Hence, the combinatorial use of a subset of the approximately 2000 human transcription factors easily accounts for the unique regulation of each gene in the human genome during development
.
or promoter regions of DNA adjacent to the genes that they regulate. Depending on the transcription factor, the transcription of the adjacent gene is either up- or down-regulated. Transcription factors use a variety of mechanisms for the regulation of gene expression. These mechanisms include:
s, an important class of transcription factors called general transcription factor
s (GTFs) are necessary for transcription to occur. Many of these GTFs don't actually bind DNA but are part of the large transcription preinitiation complex
that interacts with RNA polymerase
directly. The most common GTFs are TFIIA, TFIIB, TFIID (see also TATA binding protein
), TFIIE, TFIIF, and TFIIH. The preinitiation complex binds to promoter regions of DNA upstream to the gene that they regulate.
regions of DNA adjacent to regulated genes. These transcription factors are critical to making sure that genes are expressed in the right cell at the right time and in the right amount, depending on the changing requirements of the organism.
s are involved in development. Responding to cues (stimuli), these transcription factors turn on/off the transcription of the appropriate genes, which, in turn, allows for changes in cell morphology
or activities needed for cell fate determination
and cellular differentiation
. The Hox transcription factor family, for example, is important for proper body pattern formation
in organisms as diverse as fruit flies to humans. Another example is the transcription factor encoded by the Sex-determining Region Y
(SRY) gene, which plays a major role in determining gender in humans.
within another receptive cell. If the signal requires upregulation or downregulation of genes in the recipient cell, often transcription factors will be downstream in the signaling cascade. Estrogen
signaling is an example of a fairly short signaling cascade that involves the estrogen receptor
transcription factor: Estrogen is secreted by tissues such as the ovaries
and placenta
, crosses the cell membrane
of the recipient cell, and is bound by the estrogen receptor in the cell's cytoplasm
. The estrogen receptor then goes to the cell's nucleus
and binds to its DNA-binding sites, changing the transcriptional regulation of the associated genes.
(HSF), which upregulates genes necessary for survival at higher temperatures, hypoxia inducible factor (HIF), which upregulates genes necessary for cell survival in low-oxygen environments, and sterol regulatory element binding protein
(SREBP), which helps maintain proper lipid
levels in the cell.
, help regulate the cell cycle
and as such determine how large a cell will get and when it can divide into two daughter cells. One example is the Myc
oncogene, which has important roles in cell growth
and apoptosis
.
s) secreted by Xanthomonas
bacteria. When injected into plants, these proteins can enter the nucleus of the plant cell, bind plant promoter sequences, and activate transcription of plant genes that aid in bacterial infection. TAL effectors contain a central repeat region in which there is a simple relationship between the identity of two critical residues in sequential repeats and sequential DNA bases in the TAL effector’s target site. This property likely makes it easier for these proteins to evolve in order to better compete with the defense mechanisms of the host cell.
s, transcription factors (like most proteins) are transcribed in the nucleus
but are then translated in the cell's cytoplasm
. Many proteins that are active in the nucleus contain nuclear localization signal
s that direct them to the nucleus. But, for many transcription factors, this is a key point in their regulation. Important classes of transcription factors such as some nuclear receptor
s must first bind a ligand
while in the cytoplasm before they can relocate to the nucleus.
s in the compact particles, the nucleosome
s, where about 147 DNA base pairs make two turns around the histone protein octamer. DNA within nucleosomes is inaccessible to many transcription factors. Some transcription factors, so-called pioneering factors are still able to bind their DNA binding sites on the nucleosomal DNA. For most of other transcription factors, the nucleosome should be actively removed by molecular motors such as chromatin remodelers
. Alternatively, the nucleosome can be partially unwrapped by thermal fluctuations allowing temporary access to the transcription factor binding site. In many cases a transcription factor needs to compete for binding
to its DNA binding site with other transcription factors and histones or non-histone chromatin proteins. Pairs of transcription factors and other proteins can play antagonistic roles (activator versus repressor) in the regulation of the same gene
.
s that allow efficient recruitment of the preinitiation complex
and RNA polymerase
. Thus, for a single transcription factor to initiate transcription, all of these other proteins must also be present, and the transcription factor must be in a state where it can bind to them if necessary.
 Transcription factors are modular in structure and contain the following domains:
Transcription factors are modular in structure and contain the following domains:
transcription factor is mediated by acidic amino acids, whereas hydrophobic residues in Gcn4 play a similar role. Hence, the TADs in Gal4 and Gcn4 are referred to as acidic or hydrophobic activation domains, respectively.
Nine-amino-acid transactivation domain (9aaTAD) defines a novel domain common to a large superfamily of eukaryotic transcription factors represented by Gal4, Oaf1, Leu3, Rtg3, Pho4
, Gln3, Gcn4 in yeast and by p53
, NFAT
, NF-κB and VP16
in mammals. Martin Piskacek, Nature Precedings http://precedings.nature.com/documents/3488/version/2 (2009);
Martin Piskacek, Nature Precedings http://precedings.nature.com/documents/3939/version/1 (2009);
Martin Piskacek, Nature Precedings http://precedings.nature.com/documents/3984/version/1 (2009) Prediction for 9aa TADs (for both acidic and hydrophilic transactivation domains) is available online from ExPASy and EMBnet Spain
9aaTAD transcription factors p53
, VP16
, MLL
, E2A
, HSF1
, NF-IL6
, NFAT1 and NF-κB interact directly with the general coactivators TAF9
and CBP/p300. p53 9aaTADs interact with TAF9, GCN5 and with multiple domains of CBP/p300 (KIX, TAZ1,TAZ2 and IBiD).
KIX domain of general coactivators Med15(Gal11) interacts with 9aaTAD transcription factors Gal4, Pdr1, Oaf1, Gcn4, VP16, Pho4, Msn2, Ino2 and P201. Interactions of Gal4, Pdr1 and Gcn4 with Taf9 were reported. 9aaTAD is a common transactivation domain recruits multiple general coactivators TAF9, MED15, CBP/p300 and GCN5.
.
Transcription factors interact with their binding sites using a combination of electrostatic
(of which hydrogen bond
s are a special case) and Van der Waals force
s. Due to the nature of these chemical interactions, most transcription factors bind DNA in a sequence specific manner. However, not all bases
in the transcription factor-binding site may actually interact with the transcription factor. In addition, some of these interactions may be weaker than others. Thus, transcription factors do not bind just one sequence but are capable of binding a subset of closely related sequences, each with a different strength of interaction.
For example, although the consensus binding site
for the TATA-binding protein (TBP) is TATAAAA, the TBP transcription factor can also bind similar sequences such as TATATAT or TATATAA.
Because transcription factors can bind a set of related sequences and these sequences tend to be short, potential transcription factor binding sites can occur by chance if the DNA sequence is long enough. It is unlikely, however, that a transcription factor binds all compatible sequences in the genome
of the cell
. Other constraints, such as DNA accessibility in the cell or availability of cofactor
s may also help dictate where a transcription factor will actually bind. Thus, given the genome sequence it is still difficult to predict where a transcription factor will actually bind in a living cell.
Additional recognition specificity, however, may be obtained through the use of more than one DNA-binding domain (for example tandem DBDs in the same transcription factor or through dimerization of two transcription factors) that bind to two or more adjacent sequences of DNA.
s in transcription factors.
Many transcription factors are either tumor suppressors or oncogene
s, and, thus, mutations or aberrant regulation of them is associated with cancer. Three groups of transcription factors are known to be important in human cancer: (1) the NF-kappaB and AP-1 families, (2) the STAT
family and (3) the steroid receptor
s.
Below are a few of the more well-studied examples:
class of transcription factors. Examples include tamoxifen
and bicalutamide
for the treatment of breast
and prostate cancer
, respectively, and various types of anti-inflammatory and anabolic
steroid
s. In addition, transcription factors are often indirectly modulated by drugs through signaling cascades. It might be possible to directly target other less-explored transcription factors such as NF-κB with drugs. Transcription factors outside the nuclear receptor family are thought to be more difficult to target with small molecule
therapeutics since it is not clear that they are "drugable" but progress has been made on the notch
pathway.
and database research are commonly used. The protein version of the transcription factor is detectable by using specific antibodies. The sample is detected on a western blot
. By using electrophoretic mobility shift assay
(EMSA), the activation profile of transcription factors can be detected. A multiplex
approach for activation profiling is a TF chip system where several of different transcription factors can be detected in parallel. This technology is based on DNA microarrays, providing the specific DNA-binding sequence for the transcription factor protein on the array surface.
of their DNA-binding domains:
Molecular biology
Molecular biology is the branch of biology that deals with the molecular basis of biological activity. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry...
and genetics
Genetics
Genetics , a discipline of biology, is the science of genes, heredity, and variation in living organisms....
, a transcription factor (sometimes called a sequence-specific DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
-binding factor) is a protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
that binds to specific DNA sequence
DNA sequence
The sequence or primary structure of a nucleic acid is the composition of atoms that make up the nucleic acid and the chemical bonds that bond those atoms. Because nucleic acids, such as DNA and RNA, are unbranched polymers, this specification is equivalent to specifying the sequence of...
s, thereby controlling the flow (or transcription
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
) of genetic information from DNA to mRNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
. Transcription factors perform this function alone or with other proteins in a complex, by promoting (as an activator
Activator (genetics)
An activator is a DNA-binding protein that regulates one or more genes by increasing the rate of transcription. The activator may increase transcription by virtue of a connected domain which assists in the formation of the RNA polymerase holoenzyme, or may operate through a coactivator. A...
), or blocking (as a repressor
Repressor
In molecular genetics, a repressor is a DNA-binding protein that regulates the expression of one or more genes by binding to the operator and blocking the attachment of RNA polymerase to the promoter, thus preventing transcription of the genes. This blocking of expression is called...
) the recruitment of RNA polymerase
RNA polymerase
RNA polymerase is an enzyme that produces RNA. In cells, RNAP is needed for constructing RNA chains from DNA genes as templates, a process called transcription. RNA polymerase enzymes are essential to life and are found in all organisms and many viruses...
(the enzyme that performs the transcription
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
of genetic information from DNA to RNA) to specific genes.
A defining feature of transcription factors is that they contain one or more DNA-binding domain
DNA-binding domain
A DNA-binding domain is an independently folded protein domain that contains at least one motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA...
s (DBDs), which attach to specific sequences of DNA adjacent to the genes that they regulate. Additional proteins such as coactivator
Coactivator (genetics)
A coactivator is a protein that increases gene expression by binding to an activator which contains a DNA binding domain. The coactivator is unable to bind DNA by itself....
s, chromatin remodeler
Chromatin Structure Remodeling (RSC) Complex
RSC is a 17-subunit complex with the capacity to remodel the structure of chromatin. It exhibits a DNA-dependent ATPase activity stimulated by both free and nucleosomal DNA and a capacity to perturb nucleosome structures...
s, histone acetylases
Histone acetyltransferase
Histone acetyltransferases are enzymes that acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl CoA to form ε-N-acetyl lysine....
, deacetylases
Histone deacetylase
Histone deacetylases are a class of enzymes that remove acetyl groups from an ε-N-acetyl lysine amino acid on a histone. This is important because DNA is wrapped around histones, and DNA expression is regulated by acetylation and de-acetylation. Its action is opposite to that of histone...
, kinase
Kinase
In chemistry and biochemistry, a kinase is a type of enzyme that transfers phosphate groups from high-energy donor molecules, such as ATP, to specific substrates, a process referred to as phosphorylation. Kinases are part of the larger family of phosphotransferases...
s, and methylase
Methylase
A methylase is an enzyme that attaches a methyl group to a molecule.These are found in prokaryotes and eukaryotes. Bacteria use methylase to differentiate between foreign genetic material and their own, thus protecting their DNA from their own immune system...
s, while also playing crucial roles in gene regulation
Regulation of gene expression
Gene modulation redirects here. For information on therapeutic regulation of gene expression, see therapeutic gene modulation.Regulation of gene expression includes the processes that cells and viruses use to regulate the way that the information in genes is turned into gene products...
, lack DNA-binding domains, and, therefore, are not classified as transcription factors.
Conservation in different organisms
Transcription factors are essential for the regulation of gene expression and are, as a consequence, found in all living organisms. The number of transcription factors found within an organism increases with genome size, and larger genomes tend to have more transcription factors per gene.There are approximately 2600 proteins in the human genome
Human genome
The human genome is the genome of Homo sapiens, which is stored on 23 chromosome pairs plus the small mitochondrial DNA. 22 of the 23 chromosomes are autosomal chromosome pairs, while the remaining pair is sex-determining...
that contain DNA-binding domains, and most of these are presumed to function as transcription factors. Therefore, approximately 10% of genes in the genome code for transcription factors, which makes this family the single largest family of human proteins. Furthermore, genes are often flanked by several binding sites for distinct transcription factors, and efficient expression of each of these genes requires the cooperative action of several different transcription factors (see, for example, hepatocyte nuclear factors). Hence, the combinatorial use of a subset of the approximately 2000 human transcription factors easily accounts for the unique regulation of each gene in the human genome during development
Developmental biology
Developmental biology is the study of the process by which organisms grow and develop. Modern developmental biology studies the genetic control of cell growth, differentiation and "morphogenesis", which is the process that gives rise to tissues, organs and anatomy.- Related fields of study...
.
Mechanism
Transcription factors bind to either enhancerEnhancer (genetics)
In genetics, an enhancer is a short region of DNA that can be bound with proteins to enhance transcription levels of genes in a gene cluster...
or promoter regions of DNA adjacent to the genes that they regulate. Depending on the transcription factor, the transcription of the adjacent gene is either up- or down-regulated. Transcription factors use a variety of mechanisms for the regulation of gene expression. These mechanisms include:
- stabilize or block the binding of RNA polymerase to DNA
- catalyze the acetylationAcetylationAcetylation describes a reaction that introduces an acetyl functional group into a chemical compound...
or deacetylation of histoneHistoneIn biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
proteins. The transcription factor can either do this directly or recruit other proteins with this catalytic activity. Many transcription factors use one or the other of two opposing mechanisms to regulate transcription:- histone acetyltransferaseHistone acetyltransferaseHistone acetyltransferases are enzymes that acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl CoA to form ε-N-acetyl lysine....
(HAT) activity – acetylates histoneHistoneIn biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
proteins, which weakens the association of DNA with histoneHistoneIn biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
s, which make the DNA more accessible to transcription, thereby up-regulating transcription - histone deacetylaseHistone deacetylaseHistone deacetylases are a class of enzymes that remove acetyl groups from an ε-N-acetyl lysine amino acid on a histone. This is important because DNA is wrapped around histones, and DNA expression is regulated by acetylation and de-acetylation. Its action is opposite to that of histone...
(HDAC) activity – deacetylates histoneHistoneIn biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
proteins, which strengthens the association of DNA with histones, which make the DNA less accessible to transcription, thereby down-regulating transcription
- histone acetyltransferase
- recruit coactivatorCoactivator (genetics)A coactivator is a protein that increases gene expression by binding to an activator which contains a DNA binding domain. The coactivator is unable to bind DNA by itself....
or corepressorCorepressor (genetics)In molecular genetics, a corepressor is a substance that inhibits the expression of genes. A corepressor downregulates the expression of genes not through direct interaction with a gene promoter , but rather indirectly through interaction with repressor proteins that in turn bind to the...
proteins to the transcription factor DNA complex
Function
Transcription factors are one of the groups of proteins that read and interpret the genetic "blueprint" in the DNA. They bind DNA and help initiate a program of increased or decreased gene transcription. As such, they are vital for many important cellular processes. Below are some of the important functions and biological roles transcription factors are involved in:Basal transcription regulation
In eukaryoteEukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
s, an important class of transcription factors called general transcription factor
General transcription factor
General transcription factors or basal transcription factors are protein transcription factors that have been shown to be important in the transcription of class II genes to mRNA templates...
s (GTFs) are necessary for transcription to occur. Many of these GTFs don't actually bind DNA but are part of the large transcription preinitiation complex
Preinitiation complex
The preinitiation complex is a large complex of proteins that is necessary for the transcription of protein-coding genes in eukaryotes...
that interacts with RNA polymerase
RNA polymerase
RNA polymerase is an enzyme that produces RNA. In cells, RNAP is needed for constructing RNA chains from DNA genes as templates, a process called transcription. RNA polymerase enzymes are essential to life and are found in all organisms and many viruses...
directly. The most common GTFs are TFIIA, TFIIB, TFIID (see also TATA binding protein
TATA Binding Protein
The TATA-binding protein is a general transcription factor that binds specifically to a DNA sequence called the TATA box. This DNA sequence is found about 35 base pairs upstream of the transcription start site in some eukaryotic gene promoters...
), TFIIE, TFIIF, and TFIIH. The preinitiation complex binds to promoter regions of DNA upstream to the gene that they regulate.
Differential enhancement of transcription
Other transcription factors differentially regulate the expression of various genes by binding to enhancerEnhancer (genetics)
In genetics, an enhancer is a short region of DNA that can be bound with proteins to enhance transcription levels of genes in a gene cluster...
regions of DNA adjacent to regulated genes. These transcription factors are critical to making sure that genes are expressed in the right cell at the right time and in the right amount, depending on the changing requirements of the organism.
Development
Many transcription factors in multicellular organismMulticellular organism
Multicellular organisms are organisms that consist of more than one cell, in contrast to single-celled organisms. Most life that can be seen with the the naked eye is multicellular, as are all animals and land plants.-Evolutionary history:Multicellularity has evolved independently dozens of times...
s are involved in development. Responding to cues (stimuli), these transcription factors turn on/off the transcription of the appropriate genes, which, in turn, allows for changes in cell morphology
Morphology (biology)
In biology, morphology is a branch of bioscience dealing with the study of the form and structure of organisms and their specific structural features....
or activities needed for cell fate determination
Cell fate determination
Within the field of developmental biology one goal is to understand how a particular cell develops into the final cell type , essentially how a cell’s fate is determined. Within an embryo, 4 processes play out at the cellular and tissue level to essentially create the final organism...
and cellular differentiation
Cellular differentiation
In developmental biology, cellular differentiation is the process by which a less specialized cell becomes a more specialized cell type. Differentiation occurs numerous times during the development of a multicellular organism as the organism changes from a simple zygote to a complex system of...
. The Hox transcription factor family, for example, is important for proper body pattern formation
Regional specification
In the field of developmental biology, regional specification is the process by which different areas are identified in the development of the early embryo. The process by which the cells become specified differs between organisms.-Cell fate determination:...
in organisms as diverse as fruit flies to humans. Another example is the transcription factor encoded by the Sex-determining Region Y
SRY
SRY is a sex-determining gene on the Y chromosome in the therians .This intronless gene encodes a transcription factor that is a member of the SOX gene family of DNA-binding proteins...
(SRY) gene, which plays a major role in determining gender in humans.
Response to intercellular signals
Cells can communicate with each other by releasing molecules that produce signaling cascadesSignal transduction
Signal transduction occurs when an extracellular signaling molecule activates a cell surface receptor. In turn, this receptor alters intracellular molecules creating a response...
within another receptive cell. If the signal requires upregulation or downregulation of genes in the recipient cell, often transcription factors will be downstream in the signaling cascade. Estrogen
Estrogen
Estrogens , oestrogens , or œstrogens, are a group of compounds named for their importance in the estrous cycle of humans and other animals. They are the primary female sex hormones. Natural estrogens are steroid hormones, while some synthetic ones are non-steroidal...
signaling is an example of a fairly short signaling cascade that involves the estrogen receptor
Estrogen receptor
Estrogen receptor refers to a group of receptors that are activated by the hormone 17β-estradiol . Two types of estrogen receptor exist: ER, which is a member of the nuclear hormone family of intracellular receptors, and the estrogen G protein-coupled receptor GPR30 , which is a G protein-coupled...
transcription factor: Estrogen is secreted by tissues such as the ovaries
Ovary
The ovary is an ovum-producing reproductive organ, often found in pairs as part of the vertebrate female reproductive system. Ovaries in anatomically female individuals are analogous to testes in anatomically male individuals, in that they are both gonads and endocrine glands.-Human anatomy:Ovaries...
and placenta
Placenta
The placenta is an organ that connects the developing fetus to the uterine wall to allow nutrient uptake, waste elimination, and gas exchange via the mother's blood supply. "True" placentas are a defining characteristic of eutherian or "placental" mammals, but are also found in some snakes and...
, crosses the cell membrane
Cell membrane
The cell membrane or plasma membrane is a biological membrane that separates the interior of all cells from the outside environment. The cell membrane is selectively permeable to ions and organic molecules and controls the movement of substances in and out of cells. It basically protects the cell...
of the recipient cell, and is bound by the estrogen receptor in the cell's cytoplasm
Cytoplasm
The cytoplasm is a small gel-like substance residing between the cell membrane holding all the cell's internal sub-structures , except for the nucleus. All the contents of the cells of prokaryote organisms are contained within the cytoplasm...
. The estrogen receptor then goes to the cell's nucleus
Cell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
and binds to its DNA-binding sites, changing the transcriptional regulation of the associated genes.
Response to environment
Not only do transcription factors act downstream of signaling cascades related to biological stimuli but they can also be downstream of signaling cascades involved in environmental stimuli. Examples include heat shock factorHeat Shock Factor
Heat shock factor , in molecular biology, is the name given to transcription factors that regulate the expression of the heat shock proteins. A typical example is the heat shock factor of Drosophila melanogaster.- Function :...
(HSF), which upregulates genes necessary for survival at higher temperatures, hypoxia inducible factor (HIF), which upregulates genes necessary for cell survival in low-oxygen environments, and sterol regulatory element binding protein
Sterol regulatory element binding protein
Sterol Regulatory Element-Binding Proteins are transcription factors that bind to the sterol regulatory element DNA sequence TCACNCCAC. Mammalian SREBPs are encoded by the genes SREBF1 and SREBF2. SREBPs belong to the basic-helix-loop-helix leucine zipper class of transcription factors...
(SREBP), which helps maintain proper lipid
Lipid
Lipids constitute a broad group of naturally occurring molecules that include fats, waxes, sterols, fat-soluble vitamins , monoglycerides, diglycerides, triglycerides, phospholipids, and others...
levels in the cell.
Cell cycle control
Many transcription factors, especially some that are proto-oncogenes or tumor suppressorsTumor suppressor gene
A tumor suppressor gene, or anti-oncogene, is a gene that protects a cell from one step on the path to cancer. When this gene is mutated to cause a loss or reduction in its function, the cell can progress to cancer, usually in combination with other genetic changes.-Two-hit hypothesis:Unlike...
, help regulate the cell cycle
Cell cycle
The cell cycle, or cell-division cycle, is the series of events that takes place in a cell leading to its division and duplication . In cells without a nucleus , the cell cycle occurs via a process termed binary fission...
and as such determine how large a cell will get and when it can divide into two daughter cells. One example is the Myc
Myc
Myc is a regulator gene that codes for a transcription factor. In the human genome, Myc is located on chromosome 8 and is believed to regulate expression of 15% of all genes through binding on Enhancer Box sequences and recruiting histone acetyltransferases...
oncogene, which has important roles in cell growth
Cell growth
The term cell growth is used in the contexts of cell development and cell division . When used in the context of cell division, it refers to growth of cell populations, where one cell grows and divides to produce two "daughter cells"...
and apoptosis
Apoptosis
Apoptosis is the process of programmed cell death that may occur in multicellular organisms. Biochemical events lead to characteristic cell changes and death. These changes include blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, and chromosomal DNA fragmentation...
.
Pathogenesis
Transcription factors can also be used to alter gene expression in a host cell to promote pathogenesis. A well studied example of this are the transcription-activator like effectors (TAL effectorTAL effector
TAL effectors are proteins secreted by Xanthomonas bacteria via their type III secretion system when they infect various plant species...
s) secreted by Xanthomonas
Xanthomonas
Xanthomonas is a genus of Proteobacteria, many of which cause plant diseases. Most varieties of Xanthomonas are available from the National Collection of Plant Pathogenic Bacteria in the United Kingdom and other international culture collections such as ICMP in New Zealand, CFBP in France, and...
bacteria. When injected into plants, these proteins can enter the nucleus of the plant cell, bind plant promoter sequences, and activate transcription of plant genes that aid in bacterial infection. TAL effectors contain a central repeat region in which there is a simple relationship between the identity of two critical residues in sequential repeats and sequential DNA bases in the TAL effector’s target site. This property likely makes it easier for these proteins to evolve in order to better compete with the defense mechanisms of the host cell.
Regulation
It is common in biology for important processes to have multiple layers of regulation and control. This is also true with transcription factors: Not only do transcription factors control the rates of transcription to regulate the amounts of gene products (RNA and protein) available to the cell but transcription factors themselves are regulated (often by other transcription factors). Below is a brief synopsis of some of the ways that the activity of transcription factors can be regulated:Synthesis
Transcription factors (like all proteins) are transcribed from a gene on a chromosome into RNA, and then the RNA is translated into protein. Any of these steps can be regulated to affect the production (and thus activity) of a transcription factor. One interesting implication of this is that transcription factors can regulate themselves. For example, in a negative feedback loop, the transcription factor acts as its own repressor: If the transcription factor protein binds the DNA of its own gene, it will down-regulate the production of more of itself. This is one mechanism to maintain low levels of a transcription factor in a cell.Nuclear localization
In eukaryoteEukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
s, transcription factors (like most proteins) are transcribed in the nucleus
Cell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
but are then translated in the cell's cytoplasm
Cytoplasm
The cytoplasm is a small gel-like substance residing between the cell membrane holding all the cell's internal sub-structures , except for the nucleus. All the contents of the cells of prokaryote organisms are contained within the cytoplasm...
. Many proteins that are active in the nucleus contain nuclear localization signal
Nuclear localization signal
A nuclear localization signal or sequence is an amino acid sequence which 'tags' a protein for import into the cell nucleus by nuclear transport. Typically, this signal consists of one or more short sequences of positively charged lysines or arginines exposed on the protein surface. Different...
s that direct them to the nucleus. But, for many transcription factors, this is a key point in their regulation. Important classes of transcription factors such as some nuclear receptor
Nuclear receptor
In the field of molecular biology, nuclear receptors are a class of proteins found within cells that are responsible for sensing steroid and thyroid hormones and certain other molecules...
s must first bind a ligand
Ligand (biochemistry)
In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. In a narrower sense, it is a signal triggering molecule, binding to a site on a target protein.The binding occurs by intermolecular forces, such as ionic bonds, hydrogen...
while in the cytoplasm before they can relocate to the nucleus.
Activation
Transcription factors may be activated (or deactivated) through their signal-sensing domain by a number of mechanisms including:- ligandLigand (biochemistry)In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. In a narrower sense, it is a signal triggering molecule, binding to a site on a target protein.The binding occurs by intermolecular forces, such as ionic bonds, hydrogen...
binding – Not only is ligand binding able to influence where a transcription factor is located within a cell but ligand binding can also affect whether the transcription factor is in an active state and capable of binding DNA or other cofactors (see, for example, nuclear receptorNuclear receptorIn the field of molecular biology, nuclear receptors are a class of proteins found within cells that are responsible for sensing steroid and thyroid hormones and certain other molecules...
s). - phosphorylationPhosphorylationPhosphorylation is the addition of a phosphate group to a protein or other organic molecule. Phosphorylation activates or deactivates many protein enzymes....
– Many transcription factors such as STAT proteinSTAT proteinThe STAT protein regulates many aspects of growth, survival and differentiation in cells...
s must be phosphorylatedPhosphorylationPhosphorylation is the addition of a phosphate group to a protein or other organic molecule. Phosphorylation activates or deactivates many protein enzymes....
before they can bind DNA. - interaction with other transcription factors (e.g., homo- or hetero-dimerProtein dimerIn biochemistry, a dimer is a macromolecular complex formed by two, usually non-covalently bound, macromolecules like proteins or nucleic acids...
ization) or coregulatoryTranscription coregulatorIn molecular biology and genetics, transcription coregulators are proteins that interact with transcription factors to either activate or repress the transcription of specific genes. Transcription coregulators that activate gene transcription are referred to as coactivators while those that...
proteins
Accessibility of DNA-binding site
In eukaryotes, the DNA is organized with the help of histoneHistone
In biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
s in the compact particles, the nucleosome
Nucleosome
Nucleosomes are the basic unit of DNA packaging in eukaryotes, consisting of a segment of DNA wound around a histone protein core. This structure is often compared to thread wrapped around a spool....
s, where about 147 DNA base pairs make two turns around the histone protein octamer. DNA within nucleosomes is inaccessible to many transcription factors. Some transcription factors, so-called pioneering factors are still able to bind their DNA binding sites on the nucleosomal DNA. For most of other transcription factors, the nucleosome should be actively removed by molecular motors such as chromatin remodelers
Chromatin remodeling
Chromatin remodeling is the enzyme-assisted movement of nucleosomes on DNA.This is performed by chromatin remodeling complexes like SWI/SNF , RSC and Imitation SWI complexes ....
. Alternatively, the nucleosome can be partially unwrapped by thermal fluctuations allowing temporary access to the transcription factor binding site. In many cases a transcription factor needs to compete for binding
Competitive inhibition
Competitive inhibition is a form of enzyme inhibition where binding of the inhibitor to the active site on the enzyme prevents binding of the substrate and vice versa.-Mechanism:...
to its DNA binding site with other transcription factors and histones or non-histone chromatin proteins. Pairs of transcription factors and other proteins can play antagonistic roles (activator versus repressor) in the regulation of the same gene
Gene
A gene is a molecular unit of heredity of a living organism. It is a name given to some stretches of DNA and RNA that code for a type of protein or for an RNA chain that has a function in the organism. Living beings depend on genes, as they specify all proteins and functional RNA chains...
.
Availability of other cofactors/transcription factors
Most transcription factors do not work alone. Often, for gene transcription to occur, a number of transcription factors must bind to DNA regulatory sequences. This collection of transcription factors, in turn, recruit intermediary proteins such as cofactorTranscription coregulator
In molecular biology and genetics, transcription coregulators are proteins that interact with transcription factors to either activate or repress the transcription of specific genes. Transcription coregulators that activate gene transcription are referred to as coactivators while those that...
s that allow efficient recruitment of the preinitiation complex
Preinitiation complex
The preinitiation complex is a large complex of proteins that is necessary for the transcription of protein-coding genes in eukaryotes...
and RNA polymerase
RNA polymerase
RNA polymerase is an enzyme that produces RNA. In cells, RNAP is needed for constructing RNA chains from DNA genes as templates, a process called transcription. RNA polymerase enzymes are essential to life and are found in all organisms and many viruses...
. Thus, for a single transcription factor to initiate transcription, all of these other proteins must also be present, and the transcription factor must be in a state where it can bind to them if necessary.
Structure

- DNA-binding domainDNA-binding domainA DNA-binding domain is an independently folded protein domain that contains at least one motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA...
(DBD), which attach to specific sequences of DNA (enhancerEnhancer (genetics)In genetics, an enhancer is a short region of DNA that can be bound with proteins to enhance transcription levels of genes in a gene cluster...
or Promoter: Necessary component for all vectors: used to drive transcription of the vector's transgene promoter sequences) adjacent to regulated genes. DNA sequences that bind transcription factors are often referred to as response elementsHormone response elementA hormone response element is a response element for hormones, a short sequence of DNA within the promoter of a gene that is able to bind a specific hormone receptor complex and therefore regulate transcription...
. - Trans-activating domain (TAD), which contain binding sites for other proteins such as transcription coregulatorTranscription coregulatorIn molecular biology and genetics, transcription coregulators are proteins that interact with transcription factors to either activate or repress the transcription of specific genes. Transcription coregulators that activate gene transcription are referred to as coactivators while those that...
s. These binding sites are frequently referred to as activation functions (AFs). - An optional signal sensing domain (SSD) (e.g., a ligand binding domain), which senses external signals and, in response, transmit these signals to the rest of the transcription complex, resulting in up- or down-regulation of gene expression. Also, the DBD and signal-sensing domains may reside on separate proteins that associate within the transcription complex to regulate gene expression.
Trans-activating domain
Trans-activating domains (TADs) are named after their amino acid composition. These amino acids are either essential for the activity or simply the most abundant in the TAD. Transactivation by the Gal4GAL4/UAS system
The GAL4-UAS system is a biochemical method used to study gene expression and function in organisms such as the fruit fly. It was developed by Andrea Brand and Norbert Perrimon in 1993 and is considered a powerful technique for studying the expression of genes...
transcription factor is mediated by acidic amino acids, whereas hydrophobic residues in Gcn4 play a similar role. Hence, the TADs in Gal4 and Gcn4 are referred to as acidic or hydrophobic activation domains, respectively.
Nine-amino-acid transactivation domain (9aaTAD) defines a novel domain common to a large superfamily of eukaryotic transcription factors represented by Gal4, Oaf1, Leu3, Rtg3, Pho4
Pho4
Pho4 is a basic-helix-loop-helix transcription factor found in S. cerevisiae and other yeasts.The Pho4 homodimer binds to DNA sequences containing the bHLH binding site 5'-CACGTG-3'. This sequence is found in the promoters of genes upregulated in response to phosphate availability such as the...
, Gln3, Gcn4 in yeast and by p53
P53
p53 , is a tumor suppressor protein that in humans is encoded by the TP53 gene. p53 is crucial in multicellular organisms, where it regulates the cell cycle and, thus, functions as a tumor suppressor that is involved in preventing cancer...
, NFAT
NFAT
Nuclear factor of activated T-cells is a general name applied to a family of transcription factors shown to be important in immune response. One or more members of the NFAT family is expressed in most cells of the immune system...
, NF-κB and VP16
Herpes simplex virus protein vmw65
Vmw65, also known as VP16 or α-TIF is a trans-acting protein that forms a complex with the host transcription factors Oct-1 and HCF to induce immediate early gene transcription in the herpes simplex viruses....
in mammals. Martin Piskacek, Nature Precedings http://precedings.nature.com/documents/3488/version/2 (2009);
Martin Piskacek, Nature Precedings http://precedings.nature.com/documents/3939/version/1 (2009);
Martin Piskacek, Nature Precedings http://precedings.nature.com/documents/3984/version/1 (2009) Prediction for 9aa TADs (for both acidic and hydrophilic transactivation domains) is available online from ExPASy and EMBnet Spain
9aaTAD transcription factors p53
P53
p53 , is a tumor suppressor protein that in humans is encoded by the TP53 gene. p53 is crucial in multicellular organisms, where it regulates the cell cycle and, thus, functions as a tumor suppressor that is involved in preventing cancer...
, VP16
Herpes simplex virus protein vmw65
Vmw65, also known as VP16 or α-TIF is a trans-acting protein that forms a complex with the host transcription factors Oct-1 and HCF to induce immediate early gene transcription in the herpes simplex viruses....
, MLL
MLL (gene)
Histone-lysine N-methyltransferase HRX is an enzyme that in humans is encoded by the MLL gene.MLL is a histone methyltransferase deemed a positive global regulator of gene transcription...
, E2A
TCF3
Transcription factor 3 , also known as TCF3, is a protein that in humans is encoded by the TCF3 gene...
, HSF1
HSF1
Heat shock factor protein 1 is a protein that in humans is encoded by the HSF1 gene.-Function:The product of this gene is a heat-shock transcription factor. Transcription of heat-shock genes is rapidly induced after temperature stress...
, NF-IL6
CEBPB
CCAAT/enhancer-binding protein beta is a protein that in humans is encoded by the CEBPB gene.- Function :The protein encoded by this intronless gene is a bZIP transcription factor that can bind as a homodimer to certain DNA regulatory regions. It can also form heterodimers with the related proteins...
, NFAT1 and NF-κB interact directly with the general coactivators TAF9
TAF9
TAF9 RNA polymerase II, TATA box binding protein -associated factor, 32kDa, also known as TAF9, is a protein which in humans is encoded by the TAF9 gene.- Function :...
and CBP/p300. p53 9aaTADs interact with TAF9, GCN5 and with multiple domains of CBP/p300 (KIX, TAZ1,TAZ2 and IBiD).
KIX domain of general coactivators Med15(Gal11) interacts with 9aaTAD transcription factors Gal4, Pdr1, Oaf1, Gcn4, VP16, Pho4, Msn2, Ino2 and P201. Interactions of Gal4, Pdr1 and Gcn4 with Taf9 were reported. 9aaTAD is a common transactivation domain recruits multiple general coactivators TAF9, MED15, CBP/p300 and GCN5.
DNA-binding domain
The portion (domain) of the transcription factor that binds DNA is called its DNA-binding domain. Below is a partial list of some of the major families of DNA-binding domains/transcription factors:| Family | InterPro InterPro InterPro is a database of protein families, domains and functional sites in which identifiable features found in known proteins can be applied to new protein sequences in order to functionally characterise them.... |
Pfam Pfam Pfam is a database of protein families that includes their annotations and multiple sequence alignments generated using hidden Markov models.- Features :For each family in Pfam one can:* Look at multiple alignments* View protein domain architectures... |
SCOP Structural Classification of Proteins The Structural Classification of Proteins database is a largely manual classification of protein structural domains based on similarities of their structures and amino acid sequences. A motivation for this classification is to determine the evolutionary relationship between proteins... |
|---|---|---|---|
| basic-helix-loop-helix Basic-helix-loop-helix A basic helix-loop-helix is a protein structural motif that characterizes a family of transcription factors.- Structure :The motif is characterized by two α-helices connected by a loop. In general, transcription factors including this domain are dimeric, each with one helix containing basic amino... |
|||
| basic-leucine zipper (bZIP BZIP domain The Basic Leucine Zipper Domain is found in many DNA binding eukaryotic proteins. One part of the domain contains a region that mediates sequence specific DNA binding properties and the leucine zipper that is required for the dimerization of two DNA binding regions. The DNA binding region... ) |
|||
| C-terminal effector domain of the bipartite response regulators | |||
| GCC box | |||
| helix-turn-helix Helix-turn-helix In proteins, the helix-turn-helix is a major structural motif capable of binding DNA. It is composed of two α helices joined by a short strand of amino acids and is found in many proteins that regulate gene expression... |
|||
| homeodomain proteins Homeodomain fold The homeodomain fold is a protein structural domain that binds DNA or RNA and is thus commonly found in transcription factors. The fold consists of a 60-amino acid helix-turn-helix structure in which three alpha helices are connected by short loop regions... - bind to homeobox Homeobox A homeobox is a DNA sequence found within genes that are involved in the regulation of patterns of anatomical development in animals, fungi and plants.- Discovery :... DNA sequences, which in turn encode other transcription factors. Homeodomain proteins play critical roles in the regulation of development Developmental biology Developmental biology is the study of the process by which organisms grow and develop. Modern developmental biology studies the genetic control of cell growth, differentiation and "morphogenesis", which is the process that gives rise to tissues, organs and anatomy.- Related fields of study... . |
|||
| lambda repressor-like | |||
| srf-like (serum response factor Serum response factor Serum response factor , also known as SRF, is a transcription factor.It is a member of the MADS box superfamily of transcription factors. This protein binds to the serum response element in the promoter region of target genes... ) |
|||
| paired box Pax genes Paired box genes are a family of tissue specific transcription factors containing a paired domain and usually a partial or complete homeodomain. An octapeptide may also be present... |
|||
| winged helix | |||
| zinc finger Zinc finger Zinc fingers are small protein structural motifs that can coordinate one or more zinc ions to help stabilize their folds. They can be classified into several different structural families and typically function as interaction modules that bind DNA, RNA, proteins, or small molecules... s |
|||
| * multi-domain Cys2His2 zinc fingers | |||
| * Zn2/Cys6 | |||
| * Zn2/Cys8 nuclear receptor Nuclear receptor In the field of molecular biology, nuclear receptors are a class of proteins found within cells that are responsible for sensing steroid and thyroid hormones and certain other molecules... zinc finger |
Response elements
The DNA sequence that a transcription factor binds to is called a transcription factor-binding site or response elementResponse element
Response elements are short sequences of DNA within a gene promoter region that are able to bind a specific transcription factor and regulate transcription of genes.-Examples:Examples of response elements include:*Hormone response element...
.
Transcription factors interact with their binding sites using a combination of electrostatic
Coulomb's law
Coulomb's law or Coulomb's inverse-square law, is a law of physics describing the electrostatic interaction between electrically charged particles. It was first published in 1785 by French physicist Charles Augustin de Coulomb and was essential to the development of the theory of electromagnetism...
(of which hydrogen bond
Hydrogen bond
A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond...
s are a special case) and Van der Waals force
Van der Waals force
In physical chemistry, the van der Waals force , named after Dutch scientist Johannes Diderik van der Waals, is the sum of the attractive or repulsive forces between molecules other than those due to covalent bonds or to the electrostatic interaction of ions with one another or with neutral...
s. Due to the nature of these chemical interactions, most transcription factors bind DNA in a sequence specific manner. However, not all bases
Base pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
in the transcription factor-binding site may actually interact with the transcription factor. In addition, some of these interactions may be weaker than others. Thus, transcription factors do not bind just one sequence but are capable of binding a subset of closely related sequences, each with a different strength of interaction.
For example, although the consensus binding site
Consensus sequence
In molecular biology and bioinformatics, consensus sequence refers to the most common nucleotide or amino acid at a particular position after multiple sequences are aligned. A consensus sequence is a way of representing the results of a multiple sequence alignment, where related sequences are...
for the TATA-binding protein (TBP) is TATAAAA, the TBP transcription factor can also bind similar sequences such as TATATAT or TATATAA.
Because transcription factors can bind a set of related sequences and these sequences tend to be short, potential transcription factor binding sites can occur by chance if the DNA sequence is long enough. It is unlikely, however, that a transcription factor binds all compatible sequences in the genome
Genome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
of the cell
Cell (biology)
The cell is the basic structural and functional unit of all known living organisms. It is the smallest unit of life that is classified as a living thing, and is often called the building block of life. The Alberts text discusses how the "cellular building blocks" move to shape developing embryos....
. Other constraints, such as DNA accessibility in the cell or availability of cofactor
Cofactor (biochemistry)
A cofactor is a non-protein chemical compound that is bound to a protein and is required for the protein's biological activity. These proteins are commonly enzymes, and cofactors can be considered "helper molecules" that assist in biochemical transformations....
s may also help dictate where a transcription factor will actually bind. Thus, given the genome sequence it is still difficult to predict where a transcription factor will actually bind in a living cell.
Additional recognition specificity, however, may be obtained through the use of more than one DNA-binding domain (for example tandem DBDs in the same transcription factor or through dimerization of two transcription factors) that bind to two or more adjacent sequences of DNA.
Clinical significance
Transcription factors are of clinical significance for at least two reasons: (1) mutations can be associated with specific diseases, and (2) they can be targets of medications.Disorders
Due to their important roles in development, intercellular signaling, and cell cycle, some human diseases have been associated with mutationMutation
In molecular biology and genetics, mutations are changes in a genomic sequence: the DNA sequence of a cell's genome or the DNA or RNA sequence of a virus. They can be defined as sudden and spontaneous changes in the cell. Mutations are caused by radiation, viruses, transposons and mutagenic...
s in transcription factors.
Many transcription factors are either tumor suppressors or oncogene
Oncogene
An oncogene is a gene that has the potential to cause cancer. In tumor cells, they are often mutated or expressed at high levels.An oncogene is a gene found in the chromosomes of tumor cells whose activation is associated with the initial and continuing conversion of normal cells into cancer...
s, and, thus, mutations or aberrant regulation of them is associated with cancer. Three groups of transcription factors are known to be important in human cancer: (1) the NF-kappaB and AP-1 families, (2) the STAT
STAT
STAT may mean:*STAT protein, the Signal Transducers and Activators of Transcription protein*Special Tertiary Admissions Test, a set of tests aimed at assessing the critical reasoning abilities of university applicants who lack other formal qualifications...
family and (3) the steroid receptor
Steroid hormone receptor
Steroid hormone receptors are found on the plasma membrane, in the cytosol and also in the nucleus of target cells. They are generally intracellular receptors and initiate signal transduction for steroid hormones which lead to changes in gene expression over a time period of hours to days...
s.
Below are a few of the more well-studied examples:
| Condition | Description | Locus |
|---|---|---|
| Rett syndrome Rett syndrome Rett syndrome is a neurodevelopmental disorder of the grey matter of the brain that almost exclusively affects females. The clinical features include small hands and feet and a deceleration of the rate of head growth . Repetitive hand movements, such as wringing and/or repeatedly putting hands into... |
Mutations in the MECP2 MECP2 MECP2 is a gene that provides instructions for making its protein product, MECP2, also referred to as MeCP2. MECP2 appears to be essential for the normal function of nerve cells. The protein seems to be particularly important for mature nerve cells, where it is present in high levels... transcription factor are associated with Rett syndrome Rett syndrome Rett syndrome is a neurodevelopmental disorder of the grey matter of the brain that almost exclusively affects females. The clinical features include small hands and feet and a deceleration of the rate of head growth . Repetitive hand movements, such as wringing and/or repeatedly putting hands into... , a neurodevelopmental disorder. |
Xq28 |
| Diabetes | A rare form of diabetes called MODY Mody Mody may refer to:* Maturity onset diabetes of the young * Hormusjee Naorojee Mody... (Maturity onset diabetes of the young) can be caused by mutations in hepatocyte nuclear factors Hepatocyte nuclear factors Hepatocyte nuclear factors are a group of phylogenetically unrelated transcription factors that regulate the transcription of a diverse group of genes into proteins... (HNFs) or insulin promoter factor-1 Pdx1 Pdx1 , also known as insulin promoter factor 1, is a transcription factor necessary for pancreatic development and β-cell maturation... (IPF1/Pdx1). |
multiple |
| Developmental verbal dyspraxia | Mutations in the FOXP2 FOXP2 Forkhead box protein P2 also known as FOXP2 is a protein that in humans is encoded by the FOXP2 gene, located on human chromosome 7 . FOXP2 orthologs have also been identified in all mammals for which complete genome data are available... transcription factor are associated with developmental verbal dyspraxia, a disease in which individuals are unable to produce the finely coordinated movements required for speech. |
7q31 |
| Autoimmune diseases | Mutations in the FOXP3 FOXP3 FOXP3 is a protein involved in immune system responses. A member of the FOX protein family, FOXP3 appears to function as a master regulator in the development and function of regulatory T cells.... transcription factor cause a rare form of autoimmune disease Autoimmune disease Autoimmune diseases arise from an overactive immune response of the body against substances and tissues normally present in the body. In other words, the body actually attacks its own cells. The immune system mistakes some part of the body as a pathogen and attacks it. This may be restricted to... called IPEX. |
Xp11.23-q13.3 |
| Li-Fraumeni syndrome Li-Fraumeni syndrome Li-Fraumeni syndrome is an extremely rare autosomal dominant hereditary disorder. It is named after Frederick Pei Li and Joseph F. Fraumeni, Jr., the American physicians who first recognized and described the syndrome. Li-Fraumeni syndrome greatly increases susceptibility to cancer... |
Caused by mutations in the tumor suppressor p53. | 17p13.1 |
| breast cancer Breast cancer Breast cancer is cancer originating from breast tissue, most commonly from the inner lining of milk ducts or the lobules that supply the ducts with milk. Cancers originating from ducts are known as ductal carcinomas; those originating from lobules are known as lobular carcinomas... |
The STAT STAT STAT may mean:*STAT protein, the Signal Transducers and Activators of Transcription protein*Special Tertiary Admissions Test, a set of tests aimed at assessing the critical reasoning abilities of university applicants who lack other formal qualifications... family is relevant to breast cancer Breast cancer Breast cancer is cancer originating from breast tissue, most commonly from the inner lining of milk ducts or the lobules that supply the ducts with milk. Cancers originating from ducts are known as ductal carcinomas; those originating from lobules are known as lobular carcinomas... . |
multiple |
| multiple cancers | The HOX family are involved in a variety of cancers. | multiple |
Potential drug targets
Approximately 10% of currently prescribed drugs directly target the nuclear receptorNuclear receptor
In the field of molecular biology, nuclear receptors are a class of proteins found within cells that are responsible for sensing steroid and thyroid hormones and certain other molecules...
class of transcription factors. Examples include tamoxifen
Tamoxifen
Tamoxifen is an antagonist of the estrogen receptor in breast tissue via its active metabolite, hydroxytamoxifen. In other tissues such as the endometrium, it behaves as an agonist, hence tamoxifen may be characterized as a mixed agonist/antagonist...
and bicalutamide
Bicalutamide
Bicalutamide is an oral non-steroidal anti-androgen used in the treatment of prostate cancer and hirsutism...
for the treatment of breast
Breast cancer
Breast cancer is cancer originating from breast tissue, most commonly from the inner lining of milk ducts or the lobules that supply the ducts with milk. Cancers originating from ducts are known as ductal carcinomas; those originating from lobules are known as lobular carcinomas...
and prostate cancer
Prostate cancer
Prostate cancer is a form of cancer that develops in the prostate, a gland in the male reproductive system. Most prostate cancers are slow growing; however, there are cases of aggressive prostate cancers. The cancer cells may metastasize from the prostate to other parts of the body, particularly...
, respectively, and various types of anti-inflammatory and anabolic
Anabolic steroid
Anabolic steroids, technically known as anabolic-androgen steroids or colloquially simply as "steroids", are drugs that mimic the effects of testosterone and dihydrotestosterone in the body. They increase protein synthesis within cells, which results in the buildup of cellular tissue ,...
steroid
Steroid
A steroid is a type of organic compound that contains a characteristic arrangement of four cycloalkane rings that are joined to each other. Examples of steroids include the dietary fat cholesterol, the sex hormones estradiol and testosterone, and the anti-inflammatory drug dexamethasone.The core...
s. In addition, transcription factors are often indirectly modulated by drugs through signaling cascades. It might be possible to directly target other less-explored transcription factors such as NF-κB with drugs. Transcription factors outside the nuclear receptor family are thought to be more difficult to target with small molecule
Small molecule
In the fields of pharmacology and biochemistry, a small molecule is a low molecular weight organic compound which is by definition not a polymer...
therapeutics since it is not clear that they are "drugable" but progress has been made on the notch
Notch
Notch may refer to:* The nock of an arrow* Notch , a Hip hop, R&B, reggae, dancehall and reggaeton artist* Notch signaling pathway, a cell signaling system present in most multicellular organisms...
pathway.
Analysis
There are different technologies available to analyze transcription factors. On the genomic level, DNA-sequencingSequencing
In genetics and biochemistry, sequencing means to determine the primary structure of an unbranched biopolymer...
and database research are commonly used. The protein version of the transcription factor is detectable by using specific antibodies. The sample is detected on a western blot
Western blot
The western blot is a widely used analytical technique used to detect specific proteins in the given sample of tissue homogenate or extract. It uses gel electrophoresis to separate native proteins by 3-D structure or denatured proteins by the length of the polypeptide...
. By using electrophoretic mobility shift assay
Electrophoretic mobility shift assay
An electrophoretic mobility shift assay or mobility shift electrophoresis, also referred as a gel shift assay, gel mobility shift assay, band shift assay, or gel retardation assay, is a common affinity electrophoresis technique used to study protein–DNA or protein–RNA interactions...
(EMSA), the activation profile of transcription factors can be detected. A multiplex
Multiplex (assay)
A multiplex assay is a type of laboratory procedure that simultaneously measures multiple analytes in a single assay. It is distinguished from procedures that measure one or a few analytes at a time...
approach for activation profiling is a TF chip system where several of different transcription factors can be detected in parallel. This technology is based on DNA microarrays, providing the specific DNA-binding sequence for the transcription factor protein on the array surface.
Classes
As described in more detail below, transcription factors may be classified by their (1) mechanism of action, (2) regulatory function, or (3) sequence homology (and hence structural similarity) in their DNA-binding domains.Mechanistic
There are three mechanistic classes of transcription factors:- General transcription factorGeneral transcription factorGeneral transcription factors or basal transcription factors are protein transcription factors that have been shown to be important in the transcription of class II genes to mRNA templates...
s are involved in the formation of a preinitiation complexPreinitiation complexThe preinitiation complex is a large complex of proteins that is necessary for the transcription of protein-coding genes in eukaryotes...
. The most common are abbreviated as TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH. They are ubiquitous and interact with the core promoter region surrounding the transcription start site(s) of all class II geneClass II geneA class II gene is a type of gene that codes for a protein. Class II genes are transcribed by RNAP II.Class II genes have a promoter that often contains a TATA box.Basal transcription of class II genes requires the formation of a preinitiation complex....
s. - Upstream transcription factors are proteins that bind somewhere upstream of the initiation site to stimulate or repress transcription. These are roughly synonymous with specific transcription factors, because they vary considerably depending on what recognition sequenceRecognition sequenceThe recognition sequence, sometimes also referred to as recognition site, of any DNA-binding protein motif that exhibits binding specificity, refers to the DNA sequence , to which the domain is specific...
s are present in the proximity of the gene.
| Examples of specific transcription factors | |||
|---|---|---|---|
| Factor | Structural type | Recognition sequence Recognition sequence The recognition sequence, sometimes also referred to as recognition site, of any DNA-binding protein motif that exhibits binding specificity, refers to the DNA sequence , to which the domain is specific... | Binds as |
| SP1 | Zinc finger Zinc finger Zinc fingers are small protein structural motifs that can coordinate one or more zinc ions to help stabilize their folds. They can be classified into several different structural families and typically function as interaction modules that bind DNA, RNA, proteins, or small molecules... |
5'-GGGCGG-3' | Monomer |
| AP-1 | Basic zipper | 5'-TGA(G/C)TCA-3' | Dimer |
| C/EBP Ccaat-enhancer-binding proteins CCAAT-enhancer-binding proteins are a family of transcription factors, composed of six members called C/EBP α to C/EBP ζ. They promote the expression of certain genes through interaction with their promoter. Once bound to DNA, C/EBPs can recruit so-called coactivators CCAAT-enhancer-binding... |
Basic zipper | 5'-ATTGCGCAAT-3' | Dimer |
| Heat shock factor Heat Shock Factor Heat shock factor , in molecular biology, is the name given to transcription factors that regulate the expression of the heat shock proteins. A typical example is the heat shock factor of Drosophila melanogaster.- Function :... |
Basic zipper | 5'-XGAAX-3' | Trimer |
| ATF/CREB ATF/CREB ATF-CREB is a family of transcription factors, consisting of the ATF class, as well CREB and its subtypes.... |
Basic zipper | 5'-TGACGTCA-3' | Dimer |
| c-Myc Myc Myc is a regulator gene that codes for a transcription factor. In the human genome, Myc is located on chromosome 8 and is believed to regulate expression of 15% of all genes through binding on Enhancer Box sequences and recruiting histone acetyltransferases... |
Basic-helix-loop-helix Basic-helix-loop-helix A basic helix-loop-helix is a protein structural motif that characterizes a family of transcription factors.- Structure :The motif is characterized by two α-helices connected by a loop. In general, transcription factors including this domain are dimeric, each with one helix containing basic amino... |
5'-CACGTG-3' | Dimer |
| Oct-1 POU2F1 POU domain, class 2, transcription factor 1 is a protein that in humans is encoded by the POU2F1 gene.-Interactions:POU2F1 has been shown to interact with SNAPC4, Ku80, Glucocorticoid receptor, Sp1 transcription factor, NPAT, POU2AF1, Host cell factor C1, TATA binding protein, RELA, Nuclear... |
Helix-turn-helix Helix-turn-helix In proteins, the helix-turn-helix is a major structural motif capable of binding DNA. It is composed of two α helices joined by a short strand of amino acids and is found in many proteins that regulate gene expression... |
5'-ATGCAAAT-3' | Monomer |
| NF-1 | Novel | 5'-TTGGCXXXXXGCCAA-3' | Dimer |
(G/C) = G or C X = A Adenine Adenine is a nucleobase with a variety of roles in biochemistry including cellular respiration, in the form of both the energy-rich adenosine triphosphate and the cofactors nicotinamide adenine dinucleotide and flavin adenine dinucleotide , and protein synthesis, as a chemical component of DNA... , T Thymine Thymine is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine nucleobase. As the name suggests, thymine may be derived by methylation of uracil at... , G Guanine Guanine is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine . In DNA, guanine is paired with cytosine. With the formula C5H5N5O, guanine is a derivative of purine, consisting of a fused pyrimidine-imidazole ring system with... or C Cytosine Cytosine is one of the four main bases found in DNA and RNA, along with adenine, guanine, and thymine . It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached . The nucleoside of cytosine is cytidine... |
|||
Functional
Transcription factors have been classified according to their regulatory function:- I. constitutively-active – present in all cells at all times – general transcription factorGeneral transcription factorGeneral transcription factors or basal transcription factors are protein transcription factors that have been shown to be important in the transcription of class II genes to mRNA templates...
s, Sp1, NF1NF1NF1 can refer to:* Neurofibromatosis type I, a genetic disorder* Neurofibromin 1, a protein associated with the disorder above* Nuclear factor 1, a transcription factor...
, CCAATCcaat-enhancer-binding proteinsCCAAT-enhancer-binding proteins are a family of transcription factors, composed of six members called C/EBP α to C/EBP ζ. They promote the expression of certain genes through interaction with their promoter. Once bound to DNA, C/EBPs can recruit so-called coactivators CCAAT-enhancer-binding... - II. conditionally-active – requires activation
- II.A developmental (cell specific) – expression is tightly controlled, but, once expressed, require no additional activation – GATAGATA transcription factorGATA transcription factors are a family of transcription factors characterized by their ability to bind to the DNA sequence "GATA".-Genes:* GATA1 * GATA2 * GATA3 * GATA4 * GATA5 * GATA6...
, HNFHepatocyte nuclear factorsHepatocyte nuclear factors are a group of phylogenetically unrelated transcription factors that regulate the transcription of a diverse group of genes into proteins...
, PIT-1, MyoDMyoDMyoD is a protein with a key role in regulating muscle differentiation. MyoD belongs to a family of proteins known as myogenic regulatory factors . These bHLH transcription factors act sequentially in myogenic differentiation. MRF family members include MyoD, Myf5, myogenin, and MRF4 .MyoD is one...
, Myf5Myf5Myf5 is a protein with a key role in regulating muscle differentiation.Myf5 belongs to a family of proteins known as myogenic regulatory factors . These bHLH transcription factors act sequentially in myogenic differentiation. MRF family members include Myf5, MyoD , myogenin, and MRF4 ....
, Hox, Winged HelixWinged-helix transcription factorsConsisting of about 110 amino acids, the domain in winged-helix transcription factors has four helices and a two-strand beta-sheet.These proteins are classified into 17 families called FoxA-FoxQ.... - II.B signal-dependent – requires external signal for activation
- II.B.1 extracellular ligand (endocrineEndocrine systemIn physiology, the endocrine system is a system of glands, each of which secretes a type of hormone directly into the bloodstream to regulate the body. The endocrine system is in contrast to the exocrine system, which secretes its chemicals using ducts. It derives from the Greek words "endo"...
or paracrineParacrine signallingParacrine signalling is a form of cell signalling in which the target cell is near the signal-releasing cell.-Local action:Some signalling molecules degrade very quickly, limiting the scope of their effectiveness to the immediate surroundings...
)-dependent – nuclear receptorNuclear receptorIn the field of molecular biology, nuclear receptors are a class of proteins found within cells that are responsible for sensing steroid and thyroid hormones and certain other molecules...
s - II.B.2 intracellular ligand (autocrineAutocrine signallingAutocrine signaling is a form of signalling in which a cell secretes a hormone or chemical messenger that binds to autocrine receptors on the same cell, leading to changes in the cell...
)-dependent - activated by small intracellular molecules – SREBPSterol regulatory element binding proteinSterol Regulatory Element-Binding Proteins are transcription factors that bind to the sterol regulatory element DNA sequence TCACNCCAC. Mammalian SREBPs are encoded by the genes SREBF1 and SREBF2. SREBPs belong to the basic-helix-loop-helix leucine zipper class of transcription factors...
, p53P53p53 , is a tumor suppressor protein that in humans is encoded by the TP53 gene. p53 is crucial in multicellular organisms, where it regulates the cell cycle and, thus, functions as a tumor suppressor that is involved in preventing cancer...
, orphan nuclear receptors - II.B.3 cell membrane receptor-dependent – second messenger signaling cascades resulting in the phosphorylation of the transcription factor
- II.B.3.a resident nuclear factors – reside in the nucleus regardless of activation state – CREBCREBCREB is a cellular transcription factor. It binds to certain DNA sequences called cAMP response elements , thereby increasing or decreasing the transcription of the downstream genes....
, AP-1AP-1 (transcription factor)In the field of molecular biology, the activator protein 1 is a transcription factor which is a heterodimeric protein composed of proteins belonging to the c-Fos, c-Jun, ATF and JDP families. It regulates gene expression in response to a variety of stimuli, including cytokines, growth factors,...
, Mef2Mef2In the field of molecular biology, myocyte enhancer factor-2 proteins are a family of transcription factors which through control of gene expression are important regulators of cellular differentiation and consequently play a critical role in embryonic development. In adult organisms, Mef2... - II.B.3.b latent cytoplasmic factors – inactive form reside in the cytoplasm, but, when activated, are translocated into the nucleus – STATSTAT proteinThe STAT protein regulates many aspects of growth, survival and differentiation in cells...
, R-SMADR-SMADR-SMAD stands for receptor-regulated SMAD. Smads are transcription factors that transduce extracellular TGF-ß superfamily ligand signaling from cell membrane bound TGF-ß receptors into the nucleus where they activate transcription TGF-ß target genes...
, NF-κB, NotchNotch signalingThe notch signaling pathway is a highly conserved cell signaling system present in most multicellular organisms.Notch is present in all metazoans, and mammals possess four different notch receptors, referred to as NOTCH1, NOTCH2, NOTCH3, and NOTCH4. The notch receptor is a single-pass...
, TUBBYTubby proteinThe tubby protein is encoded by the TUB gene. It is an upstream cell signaling protein common to multicellular eukaryotes. The first tubby gene was identified in mice, and proteins that are homologous to tubby are known as "tubby-like proteins"...
, NFATNFATNuclear factor of activated T-cells is a general name applied to a family of transcription factors shown to be important in immune response. One or more members of the NFAT family is expressed in most cells of the immune system...
- II.B.3.a resident nuclear factors – reside in the nucleus regardless of activation state – CREB
- II.B.1 extracellular ligand (endocrine
- II.A developmental (cell specific) – expression is tightly controlled, but, once expressed, require no additional activation – GATA
Structural
Transcription factors are often classified based on the sequence similarity and hence the tertiary structureTertiary structure
In biochemistry and molecular biology, the tertiary structure of a protein or any other macromolecule is its three-dimensional structure, as defined by the atomic coordinates.-Relationship to primary structure:...
of their DNA-binding domains:
- 1 Superclass: Basic Domains (Basic-helix-loop-helixBasic-helix-loop-helixA basic helix-loop-helix is a protein structural motif that characterizes a family of transcription factors.- Structure :The motif is characterized by two α-helices connected by a loop. In general, transcription factors including this domain are dimeric, each with one helix containing basic amino...
)- 1.1 Class: Leucine zipperLeucine zipperA leucine zipper, aka leucine scissors, is a common three-dimensional structural motif in proteins. These motifs are usually found as part of a DNA-binding domain in various transcription factors, and are therefore involved in regulating gene expression...
factors (bZIP)- 1.1.1 Family: AP-1AP-1 (transcription factor)In the field of molecular biology, the activator protein 1 is a transcription factor which is a heterodimeric protein composed of proteins belonging to the c-Fos, c-Jun, ATF and JDP families. It regulates gene expression in response to a variety of stimuli, including cytokines, growth factors,...
(-like) components; includes (c-FosC-FosIn the field of molecular biology and Genetics, c-Fos is a protein encoded by the FOS gene.-Structure and function:c-Fos is a cellular proto-oncogene belonging to the immediate early gene family of transcription factors. c-Fos has a leucine-zipper DNA binding domain, and a transactivation domain at...
/c-JunC-junc-Jun is the name of a gene and protein that, in combination with c-Fos, forms the AP-1 early response transcription factor. It was first identified as the Fos-binding protein p39 and only later rediscovered as the product of the c-jun gene. It is activated through double phosphorylation by the...
) - 1.1.2 Family: CREBCREBCREB is a cellular transcription factor. It binds to certain DNA sequences called cAMP response elements , thereby increasing or decreasing the transcription of the downstream genes....
- 1.1.3 Family: C/EBPCcaat-enhancer-binding proteinsCCAAT-enhancer-binding proteins are a family of transcription factors, composed of six members called C/EBP α to C/EBP ζ. They promote the expression of certain genes through interaction with their promoter. Once bound to DNA, C/EBPs can recruit so-called coactivators CCAAT-enhancer-binding...
-like factors - 1.1.4 Family: bZIP / PAR
- 1.1.5 Family: Plant G-box binding factors
- 1.1.6 Family: ZIP only
- 1.1.1 Family: AP-1
- 1.2 Class: Helix-loop-helix factors (bHLH)
- 1.2.1 Family: Ubiquitous (class A) factors
- 1.2.2 Family: Myogenic transcription factors (MyoDMyoDMyoD is a protein with a key role in regulating muscle differentiation. MyoD belongs to a family of proteins known as myogenic regulatory factors . These bHLH transcription factors act sequentially in myogenic differentiation. MRF family members include MyoD, Myf5, myogenin, and MRF4 .MyoD is one...
) - 1.2.3 Family: Achaete-Scute
- 1.2.4 Family: Tal/Twist/Atonal/Hen
- 1.3 Class: Helix-loop-helix / leucine zipper factors (bHLH-ZIPBasic helix-loop-helix leucine zipper transcription factorsBasic helix-loop-helix leucine zipper transcription factors are, as their name indicates, transcription factors containing both Basic helix-loop-helix and leucine zipper motifs.An example is Microphthalmia-associated transcription factor....
)- 1.3.1 Family: Ubiquitous bHLH-ZIP factors; includes USF (USF1USF1Upstream stimulatory factor 1 is a protein that in humans is encoded by the USF1 gene.-Interactions:USF1 has been shown to interact with USF2, FOSL1 and GTF2I.-External links:...
, USF2USF2Upstream stimulatory factor 2 is a protein that in humans is encoded by the USF2 gene.-Interactions:USF2 has been shown to interact with USF1 , PPRC1 and BRCA1.- Regulation :The USF2 gene is repressed by the microRNA miR-10a....
); SREBP (SREBPSterol regulatory element binding proteinSterol Regulatory Element-Binding Proteins are transcription factors that bind to the sterol regulatory element DNA sequence TCACNCCAC. Mammalian SREBPs are encoded by the genes SREBF1 and SREBF2. SREBPs belong to the basic-helix-loop-helix leucine zipper class of transcription factors...
) - 1.3.2 Family: Cell-cycle controlling factors; includes c-MycMycMyc is a regulator gene that codes for a transcription factor. In the human genome, Myc is located on chromosome 8 and is believed to regulate expression of 15% of all genes through binding on Enhancer Box sequences and recruiting histone acetyltransferases...
- 1.3.1 Family: Ubiquitous bHLH-ZIP factors; includes USF (USF1
- 1.4 Class: NF-1
- 1.4.1 Family: NF-1 (ANFIANuclear factor 1 A-type is a protein that in humans is encoded by the NFIA gene....
, BNFIB (gene)Nuclear factor 1 B-type is a protein that in humans is encoded by the NFIB gene....
, CNFIC (gene)Nuclear factor 1 C-type is a protein that in humans is encoded by the NFIC gene....
, XNFIXNuclear factor 1 X-type is a protein that in humans is encoded by the NFIX gene.-Interactions:NFIX has been shown to interact with SKI protein....
)
- 1.4.1 Family: NF-1 (A
- 1.5 Class: RF-X
- 1.5.1 Family: RF-X (1RFX1MHC class II regulatory factor RFX1 is a protein that in humans is encoded by the RFX1 gene.-Further reading:...
, 2RFX2DNA-binding protein RFX2 is a protein that in humans is encoded by the RFX2 gene....
, 3RFX3Transcription factor RFX3 is a protein that in humans is encoded by the RFX3 gene.-Further reading:...
, 4RFX4Transcription factor RFX4 is a protein that in humans is encoded by the RFX4 gene....
, 5RFX5DNA-binding protein RFX5 is a protein that in humans is encoded by the RFX5 gene....
, ANKRFXANKDNA-binding protein RFXANK is a protein that in humans is encoded by the RFXANK gene.-Interactions:RFXANK has been shown to interact with RFXAP and CIITA.-Further reading:...
)
- 1.5.1 Family: RF-X (1
- 1.6 Class: bHSH
- 1.1 Class: Leucine zipper
- 2 Superclass: Zinc-coordinating DNA-binding domains
- 2.1 Class: Cys4 zinc fingerZinc fingerZinc fingers are small protein structural motifs that can coordinate one or more zinc ions to help stabilize their folds. They can be classified into several different structural families and typically function as interaction modules that bind DNA, RNA, proteins, or small molecules...
of nuclear receptorNuclear receptorIn the field of molecular biology, nuclear receptors are a class of proteins found within cells that are responsible for sensing steroid and thyroid hormones and certain other molecules...
type- 2.1.1 Family: Steroid hormone receptorSteroid hormone receptorSteroid hormone receptors are found on the plasma membrane, in the cytosol and also in the nucleus of target cells. They are generally intracellular receptors and initiate signal transduction for steroid hormones which lead to changes in gene expression over a time period of hours to days...
s - 2.1.2 Family: Thyroid hormone receptorThyroid hormone receptorThe thyroid hormone receptor is a type of nuclear receptor that is activated by binding thyroid hormone.-Function:Amongst the most important functions of thyroid hormone receptors are regulation of metabolism and heart rate...
-like factors
- 2.1.1 Family: Steroid hormone receptor
- 2.2 Class: diverse Cys4 zinc fingers
- 2.2.1 Family: GATA-FactorsGATA transcription factorGATA transcription factors are a family of transcription factors characterized by their ability to bind to the DNA sequence "GATA".-Genes:* GATA1 * GATA2 * GATA3 * GATA4 * GATA5 * GATA6...
- 2.2.1 Family: GATA-Factors
- 2.3 Class: Cys2His2 zinc finger domain
- 2.3.1 Family: Ubiquitous factors, includes TFIIIA, Sp1
- 2.3.2 Family: Developmental / cell cycle regulators; includes KrüppelKrüppelKrüppel is a gap gene originally described in Drosophila melanogaster, which encodes a zinc finger transcription factor with four tandemly repeated zinc finger domains....
- 2.3.4 Family: Large factors with NF-6B-like binding properties
- 2.4 Class: Cys6 cysteine-zinc cluster
- 2.5 Class: Zinc fingers of alternating composition
- 2.1 Class: Cys4 zinc finger
- 3 Superclass: Helix-turn-helixHelix-turn-helixIn proteins, the helix-turn-helix is a major structural motif capable of binding DNA. It is composed of two α helices joined by a short strand of amino acids and is found in many proteins that regulate gene expression...
- 3.1 Class: Homeo domainHomeoboxA homeobox is a DNA sequence found within genes that are involved in the regulation of patterns of anatomical development in animals, fungi and plants.- Discovery :...
- 3.1.1 Family: Homeo domain only; includes Ubx
- 3.1.2 Family: POU domain factors; includes OctOctamer transcription factorAn octamer transcription factor is a transcription factor which binds to the "ATTTGCAT" sequence.Examples include:* Oct-1 - * Oct-2 - * Oct-3/4 – * Oct-6 – * Oct-7 – * Oct-8 - * Oct-9 – * Oct-11 –...
- 3.1.3 Family: Homeo domain with LIM region
- 3.1.4 Family: homeo domain plus zinc finger motifs
- 3.2 Class: Paired box
- 3.2.1 Family: Paired plus homeo domain
- 3.2.2 Family: Paired domain only
- 3.3 Class: Fork headFOX proteinsFOX proteins are a family of transcription factors that play important roles in regulating the expression of genes involved in cell growth, proliferation, differentiation, and longevity...
/ winged helixWinged-helix transcription factorsConsisting of about 110 amino acids, the domain in winged-helix transcription factors has four helices and a two-strand beta-sheet.These proteins are classified into 17 families called FoxA-FoxQ....
- 3.3.1 Family: Developmental regulators; includes forkhead
- 3.3.2 Family: Tissue-specific regulators
- 3.3.3 Family: Cell-cycle controlling factors
- 3.3.0 Family: Other regulators
- 3.4 Class: Heat Shock FactorHeat Shock FactorHeat shock factor , in molecular biology, is the name given to transcription factors that regulate the expression of the heat shock proteins. A typical example is the heat shock factor of Drosophila melanogaster.- Function :...
s- 3.4.1 Family: HSF
- 3.5 Class: Tryptophan clusters
- 3.5.1 Family: Myb
- 3.5.2 Family: Ets-type
- 3.5.3 Family: Interferon regulatory factorsInterferon regulatory factorsInterferon regulatory factors are proteins which regulate transcription of interferons .They are used in the JAK-STAT signaling pathway....
- 3.6 Class: TEA ( transcriptional enhancer factor) domain
- 3.6.1 Family: TEA (TEAD1TEAD1Transcriptional enhancer factor TEF-1 is a protein that in humans is encoded by the TEAD1 gene.-Interactions:TEAD1 has been shown to interact with MEF2C, Serum response factor and MAX.-Further reading:...
, TEAD2TEAD2Transcriptional enhancer factor TEF-4 also known as TEA domain family member 2 is a protein that in humans is encoded by the TEAD2 gene. TEAD-2 is a transcription factor.- Clinical significance :...
, TEAD3TEAD3Transcriptional enhancer factor TEF-5 is a protein that in humans is encoded by the TEAD3 gene.-Further reading:-External links:...
, TEAD4TEAD4Transcriptional enhancer factor TEF-3 is a protein that in humans is encoded by the TEAD4 gene.-Further reading:...
)
- 3.6.1 Family: TEA (TEAD1
- 3.1 Class: Homeo domain
- 4 Superclass: beta-Scaffold Factors with Minor Groove Contacts
- 4.1 Class: RHR (Rel homology regionRel homology domainThe Rel homology domain is a protein domain found in a family of eukaryotic transcription factors, which includes NF-κB, NFAT, among others. Some of these transcription factors appear to form multi-protein DNA-bound complexes...
)- 4.1.1 Family: Rel/ankyrinAnkyrin repeatThe ankyrin repeat is a 33-residue motif in proteins consisting of two alpha helices separated by loops, first discovered in signaling proteins in yeast Cdc10 and Drosophila Notch. Ankyrin repeats mediate protein–protein interactions and are among the most common structural motifs in known proteins...
; NF-kappaB - 4.1.2 Family: ankyrin only
- 4.1.3 Family: NFATNFATNuclear factor of activated T-cells is a general name applied to a family of transcription factors shown to be important in immune response. One or more members of the NFAT family is expressed in most cells of the immune system...
(Nuclear Factor of Activated T-cells) (NFATC1NFATC1Nuclear factor of activated T-cells, cytoplasmic 1 is a protein that in humans is encoded by the NFATC1 gene.-Further reading:- External links :...
, NFATC2NFATC2Nuclear factor of activated T-cells, cytoplasmic 2 is a protein that in humans is encoded by the NFATC2 gene.NFAT transcription factors are implicated in breast cancer, more specifically in the process of cell motility at the basis of metastasis formation...
, NFATC3NFATC3Nuclear factor of activated T-cells, cytoplasmic 3 is a protein that in humans is encoded by the NFATC3 gene.- External links :...
)
- 4.1.1 Family: Rel/ankyrin
- 4.2 Class: STAT
- 4.2.1 Family: STATSTAT proteinThe STAT protein regulates many aspects of growth, survival and differentiation in cells...
- 4.2.1 Family: STAT
- 4.3 Class: p53
- 4.3.1 Family: p53P53p53 , is a tumor suppressor protein that in humans is encoded by the TP53 gene. p53 is crucial in multicellular organisms, where it regulates the cell cycle and, thus, functions as a tumor suppressor that is involved in preventing cancer...
- 4.3.1 Family: p53
- 4.4 Class: MADS boxMADS-boxThe MADS box is a conserved sequence motif found in genes which comprise the MADS-box gene family. The MADS box encodes the DNA-binding MADS domain. The MADS domain binds to DNA sequences of high similarity to the motif CC[A/T]6GG termed the CArG-box. MADS-domain proteins are generally...
- 4.4.1 Family: Regulators of differentiation; includes (Mef2Mef2In the field of molecular biology, myocyte enhancer factor-2 proteins are a family of transcription factors which through control of gene expression are important regulators of cellular differentiation and consequently play a critical role in embryonic development. In adult organisms, Mef2...
)- 4.4.2 Family: Responders to external signals, SRF (serum response factorSerum response factorSerum response factor , also known as SRF, is a transcription factor.It is a member of the MADS box superfamily of transcription factors. This protein binds to the serum response element in the promoter region of target genes...
)
- 4.4.2 Family: Responders to external signals, SRF (serum response factor
- 4.4.1 Family: Regulators of differentiation; includes (Mef2
- 4.5 Class: beta-Barrel alpha-helix transcription factors
- 4.6 Class: TATA binding proteinTATA Binding ProteinThe TATA-binding protein is a general transcription factor that binds specifically to a DNA sequence called the TATA box. This DNA sequence is found about 35 base pairs upstream of the transcription start site in some eukaryotic gene promoters...
s- 4.6.1 Family: TBP
- 4.7.1 Family: SOX genesSOX genesSOX genes encode a family of transcription factors that bind to the minor groove in DNA, and belong to a super-family of genes characterized by a homologous sequence called the HMG box. This HMG box is a DNA binding domain that is highly conserved throughout eukaryotic species...
, SRYSRYSRY is a sex-determining gene on the Y chromosome in the therians .This intronless gene encodes a transcription factor that is a member of the SOX gene family of DNA-binding proteins... - 4.7.2 Family: TCF-1 (TCF1HNF1AHNF1 homeobox A , also known as HNF1A, is a human gene.The protein encoded by this gene is a transcription factor that is highly expressed in the liver and is involved in the regulation of the expression of several liver-specific genes.-Interactions:HNF1A has been shown to interact with PCAF, Src,...
) - 4.7.3 Family: HMG2-related, SSRP1Structure specific recognition protein 1Structure specific recognition protein 1, also known as SSRP1, is a human protein.-Interactions:Structure specific recognition protein 1 has been shown to interact with NEK9....
- 4.7.5 Family: MATA
- 4.8 Class: Heteromeric CCAAT factors
- 4.8.1 Family: Heteromeric CCAAT factors
- 4.9 Class: Grainyhead
- 4.9.1 Family: Grainyhead
- 4.10 Class: Cold-shock domainCold-shock domainIm molecular biology, the cold-shock domain is a protein domain of about 70 amino acids which has been found in prokaryotic and eukaryotic DNA-binding proteins...
factors- 4.10.1 Family: csd
- 4.11 Class: Runt
- 4.11.1 Family: Runt
- 4.1 Class: RHR (Rel homology region
- 0 Superclass: Other Transcription Factors
- 0.1 Class: Copper fist proteins
- 0.2 Class: HMGI(Y) (HMGA1HMGA1High-mobility group protein HMG-I/HMG-Y is a protein that in humans is encoded by the HMGA1 gene.Mice lacking their variant of HMGA1, i.e., Hmga1-/- mice, are diabetic and express low levels of the insulin receptor.-Interactions:...
)- 0.2.1 Family: HMGI(Y)
- 0.3 Class: Pocket domain
- 0.4 Class: E1A-like factors
- 0.5 Class: AP2/EREBP-related factors
- 0.5.1 Family: AP2Apetala 2Apetala 2 ' is a gene coding for a member of a large family of transcription factors, the AP2/EREBP family. In Arabidopsis thaliana which plays a role in the ABC model of flower development...
- 0.5.2 Family: EREBP
- 0.5.3 Superfamily: AP2/B3
- 0.5.3.1 Family: ARF
- 0.5.3.2 Family: ABI
- 0.5.3.3 Family: RAV
- 0.5.1 Family: AP2
External links
- Protein-DNA binding: data, tools & models Figure 8-10 from Essential cell biology.

