
Nucleosome
Encyclopedia
Nucleosomes are the basic unit of DNA
packaging in eukaryotes, consisting of a segment of DNA wound around a histone
protein
core. This structure is often compared to thread wrapped around a spool.
Nucleosomes form the fundamental repeating units of eukaryotic
chromatin
, which is used to pack the large eukaryotic genomes into the nucleus while still ensuring appropriate access to it (in mammalian cells approximately 2 m of linear DNA
have to be packed into a nucleus
of roughly 10 µm diameter). Nucleosomes are folded through a series of successively higher order structures to eventually form a chromosome
; this both compacts DNA and creates an added layer of regulatory control, which ensures correct gene expression. Nucleosomes are thought to carry epigenetically
inherited information in the form of covalent modifications of their core histones.
The nucleosome hypothesis was proposed by Don and Ada Olins in 1974 and Roger Kornberg.
The nucleosome core particle consists of approximately 147 base pair
s of DNA
wrapped in 1.67 left-handed superhelical turns around a histone
octamer consisting of 2 copies each of the core histones H2A
, H2B
, H3
, and H4
. Core particles are connected by stretches of "linker DNA", which can be up to about 80 bp long. Technically, a nucleosome is defined as the core particle plus one of these linker regions; however the word is often synonymous with the core particle.
Linker histones such as H1 and its isoforms are involved in chromatin compaction and sit at the base of the nucleosome near the DNA entry and exit binding to the linker region of the DNA. Non-condensed nucleosomes without the linker histone resemble "beads on a string of DNA" under an electron microscope
.
In contrast to most eukaryotic cells, mature sperm cells largely use protamines to package their genomic DNA, most likely to achieve an even higher packaging ratio. Histone equivalents and a simplified chromatin structure have also been found in Archea, proving that eukaryotes are not the only organisms that use nucleosomes.
of the nucleosome was solved by the Richmond group, showing some of the most important details of the particle. The human alpha-satellite palindromic DNA critical to achieving the 1997 nucleosome crystal structure was developed by the Bunick group at Oak Ridge National Laboratory in Tennessee. The structures of over 20 different nucleosome core particles have been solved to date, including those containing histone variants and histones from different species. The structure of the nucleosome core particle is remarkably conserved, and even a change of over 100 residues between frog and yeast histones results in electron density maps with an overall root mean square deviation
of only 1.6Å.
of DNA wrapped in 1.67 left-handed superhelical turns around the histone octamer
, consisting of 2 copies each of the core histones H2A
, H2B
, H3
, and H4
. Adjacent nucleosomes are joined by a stretch of free DNA termed "linker DNA" (which varies from 10 - 80 bp in length depending on species and tissue type).
Nucleosome core particles are observed when chromatin in interphase is treated to cause the chromatin to unfold partially. The resulting image, via an electron microscope, is "beads on a string". The string is the DNA, while each bead in the nucleosome is a core particle. The nucleosome core particle is composed of DNA and histone proteins.
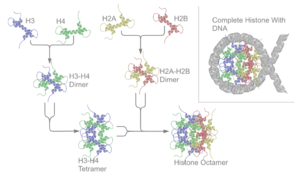
The core histone proteins contain a characteristic structural motif termed the "histone fold," which consists of three alpha-helices (α1-3) separated by two loops (L1-2). In solution, the histones form H2A-H2B heterodimers and H3-H4 heterotetramers. Histones dimerise about their long α2 helices in an anti-parallel orientation, and, in the case of H3 and H4, two such dimers form a 4-helix bundle stabilised by extensive H3-H3’ interaction. The H2A/H2B dimer binds onto the H3/H4 tetramer due to interactions between H4 and H2B, which include the formation of a hydrophobic cluster.
The histone octamer is formed by a central H3/H4 tetramer sandwiched between two H2A/H2B dimers. Due to the highly basic charge of all four core histones, the histone octamer is stable only in the presence of DNA or very high salt concentrations.
The nucleosome contains over 120 direct protein-DNA interactions and several hundred water-mediated ones. Direct protein - DNA interactions are not spread evenly about the octamer surface but rather located at discrete sites. These are due to the formation of two types of DNA binding sites within the octamer; the α1α1 site, which uses the α1 helix from two adjacent histones, and the L1L2 site formed by the L1 and L2 loops. Salt links and hydrogen bonding between both side-chain basic and hydroxyl groups and main-chain amides with the DNA backbone phosphates form the bulk of interactions with the DNA. This is important, given that the ubiquitous distribution of nucleosomes along genomes requires it to be a non-sequence-specific DNA-binding factor. Although nucleosomes tend to prefer some DNA sequences over others, they are capable of binding practically to any sequence, which is thought to be due to the flexibility in the formation of these water-mediated interactions. In addition, non-polar interactions are made between protein side-chains and the deoxyribose groups, and an arginine side-chain intercalates into the DNA minor groove at all 14 sites it faces the octamer surface.
The distribution and strength of DNA-binding sites about the octamer surface distorts the DNA within the nucleosome core. The DNA is non-uniformly bent and also contains twist defects. The twist of free B-form DNA in solution is 10.5 bp per turn. However, the overall twist of nucleosomal DNA is only 10.2 bp per turn, varying from a value of 9.4 to 10.9 bp per turn.
of the H4 tail distorts the higher-order structure of chromatin.
 The organization of the DNA that is achieved by the nucleosome cannot fully explain the packaging of DNA observed in the cell nucleus. Further compaction of chromatin
The organization of the DNA that is achieved by the nucleosome cannot fully explain the packaging of DNA observed in the cell nucleus. Further compaction of chromatin
into the cell nucleus is necessary, but is not yet well understood. The current understanding is that repeating nucleosomes with intervening "linker" DNA form a 10-nm-fiber, described as "beads on a string", and have a packing ratio of about five to ten. A chain of nucleosomes can be arranged in a 30 nm fiber, a compacted structure with a packing ratio of ~50 and whose formation is dependent on the presence of the H1 histone
.
A crystal structure of a tetranucleosome has been presented and used to build up a proposed structure of the 30 nm fiber as a two-start helix.
There is still a certain amount of contention regarding this model, as it is incompatible with recent electron microscopy data. Beyond this, the structure of chromatin is poorly understood, but it is classically suggested that the 30 nm fiber is arranged into loops along a central protein scaffold to form transcriptionally active euchromatin
. Further compaction leads to transcriptionally inactive heterochromatin
.
revealed that DNA within the nucleosome remains fully wrapped for only 250 ms before it is unwrapped for 10-50 ms and then rapidly rewrapped. This implies that DNA does not need to be actively dissociated from the nucleosome but that there is a significant fraction of time during which it is fully accessible. Indeed, this can be extended to the observation that introducing a DNA-binding sequence within the nucleosome increases the accessibility of adjacent regions of DNA when bound. This propensity for DNA within the nucleosome to “breathe” is predicted to have important functional consequences for all DNA-binding proteins that operate in a chromatin environment.
Some modifications have been shown to be correlated with gene silencing; others seem to be correlated with gene activation. Common modifications include acetylation
, methylation
, or ubiquitination of lysine
; methylation
of arginine
; and phosphorylation
of serine
. The information stored in this way is considered epigenetic, since it is not encoded in the DNA but is still inherited to daughter cells. The maintenance of a repressed or activated status of a gene is often necessary for cellular differentiation
.
differentiation), whereas the inactive X chromosomes
in mammals are enriched in macroH2A. H3 can be replaced by H3.3 (which correlates with activated genes) and in centromere
s H3 is replaced by CENPA
.
Studies looking at gene activation in vivo and, more astonishingly, remodelling in vitro have revealed that chromatin remodelling events and transcription-factor binding are cyclical and periodic in nature. While the consequences of this for the reaction mechanism of chromatin remodelling are not known, the dynamic nature of the system may allow it to respond faster to external stimuli.
.
About 80% of the yeast genome appears to be covered by nucleosomes and the pattern of nucleosome positioning clearly relates to DNA regions that regulate transcription
, regions that are transcribed and regions that initiate DNA replication. Most recently, a new study examined ‘’dynamic changes’’ in nucleosome repositioning during a global transcriptional reprogramming event to elucidate the effects on nucleosome displacement during genome-wide transcriptional changes in yeast (Saccharomyces cerevisiae
). The results suggested that nucleosomes that were localized to promoter regions are displaced in response to stress (like heat shock
). In addition, the removal of nucleosomes usually corresponded to transcriptional activation and the replacement of nucleosomes usually corresponded to transcriptional repression, presumably because transcription factor
binding sites became more or less accessible, respectively. In general, only one or two nucleosomes were repositioned at the promoter to effect these transcriptional changes. However, even in chromosomal regions that were not associated with transcriptional changes, nucleosome repositioning was observed, suggesting that the covering and uncovering of transcriptional DNA does not necessarily produce a transcriptional event.
 Nucleosomes can be assembled in vitro
Nucleosomes can be assembled in vitro
by either using purified native or recombinant histones. One standard technique of loading the DNA around the histones involves the use of salt dialysis
. A reaction consisting of the histone octamers and a naked DNA template can be incubated together at a salt concentration of 2 M. By steadily decreasing the salt concentration, the DNA will equilibrate to a position where it is wrapped around the histone octamers, forming nucleosomes. In appropriate conditions, this reconstitution process allows for the nucleosome positioning affinity of a given sequence to be mapped experimentally.
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
packaging in eukaryotes, consisting of a segment of DNA wound around a histone
Histone
In biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
core. This structure is often compared to thread wrapped around a spool.
Nucleosomes form the fundamental repeating units of eukaryotic
Eukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
chromatin
Chromatin
Chromatin is the combination of DNA and proteins that make up the contents of the nucleus of a cell. The primary functions of chromatin are; to package DNA into a smaller volume to fit in the cell, to strengthen the DNA to allow mitosis and meiosis and prevent DNA damage, and to control gene...
, which is used to pack the large eukaryotic genomes into the nucleus while still ensuring appropriate access to it (in mammalian cells approximately 2 m of linear DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
have to be packed into a nucleus
Cell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
of roughly 10 µm diameter). Nucleosomes are folded through a series of successively higher order structures to eventually form a chromosome
Chromosome
A chromosome is an organized structure of DNA and protein found in cells. It is a single piece of coiled DNA containing many genes, regulatory elements and other nucleotide sequences. Chromosomes also contain DNA-bound proteins, which serve to package the DNA and control its functions.Chromosomes...
; this both compacts DNA and creates an added layer of regulatory control, which ensures correct gene expression. Nucleosomes are thought to carry epigenetically
Epigenetics
In biology, and specifically genetics, epigenetics is the study of heritable changes in gene expression or cellular phenotype caused by mechanisms other than changes in the underlying DNA sequence – hence the name epi- -genetics...
inherited information in the form of covalent modifications of their core histones.
The nucleosome hypothesis was proposed by Don and Ada Olins in 1974 and Roger Kornberg.
The nucleosome core particle consists of approximately 147 base pair
Base pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
s of DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
wrapped in 1.67 left-handed superhelical turns around a histone
Histone
In biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
octamer consisting of 2 copies each of the core histones H2A
Histone H2A
Histone H2A is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N terminal tail, H2A is involved with the structure of the nucleosomes of the 'beads on a string' structure.Other histone proteins...
, H2B
Histone H2B
Histone H2B is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N terminal tail H2B is involved with the structure of the nucleosomes of the 'beads on a string' structure.See nucleosome, histone and...
, H3
Histone H3
Histone H3 is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a string' structure...
, and H4
Histone H4
Histone H4 is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N terminal tail, H4 is a structural component of the nucleosome, and is subject to covalent modification, including acetylation and methylation,...
. Core particles are connected by stretches of "linker DNA", which can be up to about 80 bp long. Technically, a nucleosome is defined as the core particle plus one of these linker regions; however the word is often synonymous with the core particle.
Linker histones such as H1 and its isoforms are involved in chromatin compaction and sit at the base of the nucleosome near the DNA entry and exit binding to the linker region of the DNA. Non-condensed nucleosomes without the linker histone resemble "beads on a string of DNA" under an electron microscope
Electron microscope
An electron microscope is a type of microscope that uses a beam of electrons to illuminate the specimen and produce a magnified image. Electron microscopes have a greater resolving power than a light-powered optical microscope, because electrons have wavelengths about 100,000 times shorter than...
.
In contrast to most eukaryotic cells, mature sperm cells largely use protamines to package their genomic DNA, most likely to achieve an even higher packaging ratio. Histone equivalents and a simplified chromatin structure have also been found in Archea, proving that eukaryotes are not the only organisms that use nucleosomes.
Structure of the core particle
Overview
Early structural studies provided evidence that an octamer of histone proteins wraps DNA around itself in about two turns of a left-handed superhelix. In 1997 the first near atomic resolution crystal structureCrystal structure
In mineralogy and crystallography, crystal structure is a unique arrangement of atoms or molecules in a crystalline liquid or solid. A crystal structure is composed of a pattern, a set of atoms arranged in a particular way, and a lattice exhibiting long-range order and symmetry...
of the nucleosome was solved by the Richmond group, showing some of the most important details of the particle. The human alpha-satellite palindromic DNA critical to achieving the 1997 nucleosome crystal structure was developed by the Bunick group at Oak Ridge National Laboratory in Tennessee. The structures of over 20 different nucleosome core particles have been solved to date, including those containing histone variants and histones from different species. The structure of the nucleosome core particle is remarkably conserved, and even a change of over 100 residues between frog and yeast histones results in electron density maps with an overall root mean square deviation
Root mean square deviation
The root-mean-square deviation is the measure of the average distance between the atoms of superimposed proteins...
of only 1.6Å.
The nucleosome core particle
The nucleosome core particle (shown in the figure) consists of about 146 bpBase pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
of DNA wrapped in 1.67 left-handed superhelical turns around the histone octamer
Histone octamer
A histone octamer is an octamer of the histones found at the center of a nucleosome core particle. It consists of 2 copies of each of the four core histone proteins . The octamer assembles when a tetramer, containing two copies of both H3 and H4, complexes with two H2A/H2B dimers...
, consisting of 2 copies each of the core histones H2A
Histone H2A
Histone H2A is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N terminal tail, H2A is involved with the structure of the nucleosomes of the 'beads on a string' structure.Other histone proteins...
, H2B
Histone H2B
Histone H2B is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N terminal tail H2B is involved with the structure of the nucleosomes of the 'beads on a string' structure.See nucleosome, histone and...
, H3
Histone H3
Histone H3 is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a string' structure...
, and H4
Histone H4
Histone H4 is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N terminal tail, H4 is a structural component of the nucleosome, and is subject to covalent modification, including acetylation and methylation,...
. Adjacent nucleosomes are joined by a stretch of free DNA termed "linker DNA" (which varies from 10 - 80 bp in length depending on species and tissue type).
Nucleosome core particles are observed when chromatin in interphase is treated to cause the chromatin to unfold partially. The resulting image, via an electron microscope, is "beads on a string". The string is the DNA, while each bead in the nucleosome is a core particle. The nucleosome core particle is composed of DNA and histone proteins.
Protein interactions within the nucleosome
The core histone proteins contain a characteristic structural motif termed the "histone fold," which consists of three alpha-helices (α1-3) separated by two loops (L1-2). In solution, the histones form H2A-H2B heterodimers and H3-H4 heterotetramers. Histones dimerise about their long α2 helices in an anti-parallel orientation, and, in the case of H3 and H4, two such dimers form a 4-helix bundle stabilised by extensive H3-H3’ interaction. The H2A/H2B dimer binds onto the H3/H4 tetramer due to interactions between H4 and H2B, which include the formation of a hydrophobic cluster.
The histone octamer is formed by a central H3/H4 tetramer sandwiched between two H2A/H2B dimers. Due to the highly basic charge of all four core histones, the histone octamer is stable only in the presence of DNA or very high salt concentrations.
Histone - DNA interactions
The nucleosome contains over 120 direct protein-DNA interactions and several hundred water-mediated ones. Direct protein - DNA interactions are not spread evenly about the octamer surface but rather located at discrete sites. These are due to the formation of two types of DNA binding sites within the octamer; the α1α1 site, which uses the α1 helix from two adjacent histones, and the L1L2 site formed by the L1 and L2 loops. Salt links and hydrogen bonding between both side-chain basic and hydroxyl groups and main-chain amides with the DNA backbone phosphates form the bulk of interactions with the DNA. This is important, given that the ubiquitous distribution of nucleosomes along genomes requires it to be a non-sequence-specific DNA-binding factor. Although nucleosomes tend to prefer some DNA sequences over others, they are capable of binding practically to any sequence, which is thought to be due to the flexibility in the formation of these water-mediated interactions. In addition, non-polar interactions are made between protein side-chains and the deoxyribose groups, and an arginine side-chain intercalates into the DNA minor groove at all 14 sites it faces the octamer surface.
The distribution and strength of DNA-binding sites about the octamer surface distorts the DNA within the nucleosome core. The DNA is non-uniformly bent and also contains twist defects. The twist of free B-form DNA in solution is 10.5 bp per turn. However, the overall twist of nucleosomal DNA is only 10.2 bp per turn, varying from a value of 9.4 to 10.9 bp per turn.
Histone tail domains
The histone tail extensions constitute up to 30% by mass of histones, but are not visible in the crystal structures of nucleosomes due to their high intrinsic flexibility, and have been thought to be largely unstructured. The N-terminal tails of histones H3 and H2B pass through a channel formed by the minor grooves of the two DNA strands, protruding from the DNA every 20 bp. The N-terminal tail of histone H4, on the other hand, has a region of highly basic amino acids (16-25), which, in the crystal structure, forms an interaction with the highly acidic surface region of a H2A-H2B dimer of another nucleosome, being potentially relevant for the higher-order structure of nucleosomes. This interaction is thought to occur under physiological conditions also, and suggests that acetylationAcetylation
Acetylation describes a reaction that introduces an acetyl functional group into a chemical compound...
of the H4 tail distorts the higher-order structure of chromatin.
Higher order structure

Chromatin
Chromatin is the combination of DNA and proteins that make up the contents of the nucleus of a cell. The primary functions of chromatin are; to package DNA into a smaller volume to fit in the cell, to strengthen the DNA to allow mitosis and meiosis and prevent DNA damage, and to control gene...
into the cell nucleus is necessary, but is not yet well understood. The current understanding is that repeating nucleosomes with intervening "linker" DNA form a 10-nm-fiber, described as "beads on a string", and have a packing ratio of about five to ten. A chain of nucleosomes can be arranged in a 30 nm fiber, a compacted structure with a packing ratio of ~50 and whose formation is dependent on the presence of the H1 histone
Histone H1
Histone H1 is one of the five main histone protein families which are components of chromatin in eukaryotic cells. Though highly conserved, it is nevertheless the most variable histone in sequence across species.- Structure :...
.
A crystal structure of a tetranucleosome has been presented and used to build up a proposed structure of the 30 nm fiber as a two-start helix.
There is still a certain amount of contention regarding this model, as it is incompatible with recent electron microscopy data. Beyond this, the structure of chromatin is poorly understood, but it is classically suggested that the 30 nm fiber is arranged into loops along a central protein scaffold to form transcriptionally active euchromatin
Euchromatin
Euchromatin is a lightly packed form of chromatin that is rich in gene concentration, and is often under active transcription. Unlike heterochromatin, it is found in both cells with nuclei and cells without nuclei...
. Further compaction leads to transcriptionally inactive heterochromatin
Heterochromatin
Heterochromatin is a tightly packed form of DNA, which comes in different varieties. These varieties lie on a continuum between the two extremes of constitutive and facultative heterochromatin...
.
Nucleosome dynamics
Although the nucleosome is a very stable protein-DNA complex, it is not static and has been shown to undergo a number of different structural re-arrangements including nucleosome sliding and DNA site exposure. Depending on the context, nucleosomes can inhibit or facilitate transcription factor binding. Nucleosome positions are controlled by three major contributions: First, the intrinsic binding affinity of the histone octamer depends on the DNA sequence. Second, the nucleosome can be displaced or recruited by the competitive or cooperative binding of other protein factors. Third, the nucleosome may be actively translocated by ATP-dependent remodeling complexes.Nucleosome sliding
Work performed in the Bradbury laboratory showed that nucleosomes reconstituted onto the 5S DNA positioning sequence were able to reposition themselves translationally onto adjacent sequences when incubated thermally. Later work showed that this repositioning did not require disruption of the histone octamer but was consistent with nucleosomes being able to “slide” along the DNA in cis. In 2008, It was further revealed that CTCF binding sites act as nucleosome positioning anchors so that, when used to align various genomic signals, multiple flanking nucleosomes can be readily identified. Although nucleosomes are intrinsically mobile, eukaryotes have evolved a large family of ATP-dependent chromatin remodelling enzymes to alter chromatin structure, many of which do so via nucleosome sliding.DNA site exposure
Work from the Widom laboratory has shown that nucleosomal DNA is in equilibrium between a wrapped and unwrapped state. Measurements of these rates using time-resolved FRETFret
A fret is a raised portion on the neck of a stringed instrument, that extends generally across the full width of the neck. On most modern western instruments, frets are metal strips inserted into the fingerboard...
revealed that DNA within the nucleosome remains fully wrapped for only 250 ms before it is unwrapped for 10-50 ms and then rapidly rewrapped. This implies that DNA does not need to be actively dissociated from the nucleosome but that there is a significant fraction of time during which it is fully accessible. Indeed, this can be extended to the observation that introducing a DNA-binding sequence within the nucleosome increases the accessibility of adjacent regions of DNA when bound. This propensity for DNA within the nucleosome to “breathe” is predicted to have important functional consequences for all DNA-binding proteins that operate in a chromatin environment.
Modulating nucleosome structure
Eukaryotic genomes are ubiquitously associated into chromatin; however, cells must spatially and temporally regulate specific loci independently of bulk chromatin. In order to achieve the high level of control required to co-ordinate nuclear processes such as DNA replication, repair, and transcription, cells have developed a variety of means to locally and specifically modulate chromatin structure and function. This can involve covalent modification of histones, the incorporation of histone variants, and non-covalent remodelling by ATP-dependent remodelling enzymes.Histone post-translational modifications
Since they were discovered in the mid-1960s, histone modifications have been predicted to affect transcription. The fact that most of the early post-translational modifications found were concentrated within the tail extensions that protrude from the nucleosome core lead to two main theories regarding the mechanism of histone modification. The first of the theories suggested that they may affect electrostatic interactions between the histone tails and DNA to “loosen” chromatin structure. Later it was proposed that combinations of these modifications may create binding epitopes with which to recruit other proteins. Recently, given that more modifications have been found in the structured regions of histones, it has been put forward that these modifications may affect histone-DNA and histone-histone interactions within the nucleosome core.Some modifications have been shown to be correlated with gene silencing; others seem to be correlated with gene activation. Common modifications include acetylation
Acetylation
Acetylation describes a reaction that introduces an acetyl functional group into a chemical compound...
, methylation
Methylation
In the chemical sciences, methylation denotes the addition of a methyl group to a substrate or the substitution of an atom or group by a methyl group. Methylation is a form of alkylation with, to be specific, a methyl group, rather than a larger carbon chain, replacing a hydrogen atom...
, or ubiquitination of lysine
Lysine
Lysine is an α-amino acid with the chemical formula HO2CCH4NH2. It is an essential amino acid, which means that the human body cannot synthesize it. Its codons are AAA and AAG....
; methylation
Methylation
In the chemical sciences, methylation denotes the addition of a methyl group to a substrate or the substitution of an atom or group by a methyl group. Methylation is a form of alkylation with, to be specific, a methyl group, rather than a larger carbon chain, replacing a hydrogen atom...
of arginine
Arginine
Arginine is an α-amino acid. The L-form is one of the 20 most common natural amino acids. At the level of molecular genetics, in the structure of the messenger ribonucleic acid mRNA, CGU, CGC, CGA, CGG, AGA, and AGG, are the triplets of nucleotide bases or codons that codify for arginine during...
; and phosphorylation
Phosphorylation
Phosphorylation is the addition of a phosphate group to a protein or other organic molecule. Phosphorylation activates or deactivates many protein enzymes....
of serine
Serine
Serine is an amino acid with the formula HO2CCHCH2OH. It is one of the proteinogenic amino acids. By virtue of the hydroxyl group, serine is classified as a polar amino acid.-Occurrence and biosynthesis:...
. The information stored in this way is considered epigenetic, since it is not encoded in the DNA but is still inherited to daughter cells. The maintenance of a repressed or activated status of a gene is often necessary for cellular differentiation
Cellular differentiation
In developmental biology, cellular differentiation is the process by which a less specialized cell becomes a more specialized cell type. Differentiation occurs numerous times during the development of a multicellular organism as the organism changes from a simple zygote to a complex system of...
.
Histone variants
Although histones are remarkably conserved throughout evolution, several variant forms have been identified. It is interesting to note that this diversification of histone function is restricted to H2A and H3, with H2B and H4 being mostly invariant. H2A can be replaced by H2AZ (which leads to reduced nucleosome stability) or H2AX (which is associated with DNA repair and T cellT cell
T cells or T lymphocytes belong to a group of white blood cells known as lymphocytes, and play a central role in cell-mediated immunity. They can be distinguished from other lymphocytes, such as B cells and natural killer cells , by the presence of a T cell receptor on the cell surface. They are...
differentiation), whereas the inactive X chromosomes
X-inactivation
X-inactivation is a process by which one of the two copies of the X chromosome present in female mammals is inactivated. The inactive X chromosome is silenced by packaging into transcriptionally inactive heterochromatin...
in mammals are enriched in macroH2A. H3 can be replaced by H3.3 (which correlates with activated genes) and in centromere
Centromere
A centromere is a region of DNA typically found near the middle of a chromosome where two identical sister chromatids come closest in contact. It is involved in cell division as the point of mitotic spindle attachment...
s H3 is replaced by CENPA
CENPA
Centromere protein A, also known as CENPA, is a protein which in humans is encoded by the CENPA gene.Centromeres are the differentiated chromosomal domains that specify the mitotic behavior of chromosomes. The CENPA gene encodes a centromere protein which contains a histone H3 related histone fold...
.
ATP-dependent nucleosome remodelling
A number of distinct reactions are associated with the term ATP-dependent chromatin remodelling. Remodelling enzymes have been shown to slide nucleosomes along DNA, disrupt histone-DNA contacts to the extent of destabilising the H2A/H2B dimer and to generate negative superhelical torsion in DNA and chromatin. Recently, the Swr1 remodelling enzyme has been shown to introduce the variant histone H2A.Z into nucleosomes. At present, it is not clear if all of these represent distinct reactions or merely alternative outcomes of a common mechanism. What is shared between all, and indeed the hallmark of ATP-dependent chromatin remodelling, is that they all result in altered DNA accessibility.Studies looking at gene activation in vivo and, more astonishingly, remodelling in vitro have revealed that chromatin remodelling events and transcription-factor binding are cyclical and periodic in nature. While the consequences of this for the reaction mechanism of chromatin remodelling are not known, the dynamic nature of the system may allow it to respond faster to external stimuli.
Dynamic nucleosome remodelling across the Yeast genome
Studies in 2007 have catalogued nucleosome positions in yeast and shown that nucleosomes are depleted in promoter regions and origins of replicationOrigin of replication
The origin of replication is a particular sequence in a genome at which replication is initiated. This can either be DNA replication in living organisms such as prokaryotes and eukaryotes, or RNA replication in RNA viruses, such as double-stranded RNA viruses...
.
About 80% of the yeast genome appears to be covered by nucleosomes and the pattern of nucleosome positioning clearly relates to DNA regions that regulate transcription
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
, regions that are transcribed and regions that initiate DNA replication. Most recently, a new study examined ‘’dynamic changes’’ in nucleosome repositioning during a global transcriptional reprogramming event to elucidate the effects on nucleosome displacement during genome-wide transcriptional changes in yeast (Saccharomyces cerevisiae
Saccharomyces cerevisiae
Saccharomyces cerevisiae is a species of yeast. It is perhaps the most useful yeast, having been instrumental to baking and brewing since ancient times. It is believed that it was originally isolated from the skin of grapes...
). The results suggested that nucleosomes that were localized to promoter regions are displaced in response to stress (like heat shock
Heat shock
In biochemistry, heat shock is the effect of subjecting a cell to a higher temperature than that of the ideal body temperature of the organism from which the cell line was derived.-Heat shock response:...
). In addition, the removal of nucleosomes usually corresponded to transcriptional activation and the replacement of nucleosomes usually corresponded to transcriptional repression, presumably because transcription factor
Transcription factor
In molecular biology and genetics, a transcription factor is a protein that binds to specific DNA sequences, thereby controlling the flow of genetic information from DNA to mRNA...
binding sites became more or less accessible, respectively. In general, only one or two nucleosomes were repositioned at the promoter to effect these transcriptional changes. However, even in chromosomal regions that were not associated with transcriptional changes, nucleosome repositioning was observed, suggesting that the covering and uncovering of transcriptional DNA does not necessarily produce a transcriptional event.
Nucleosome assembly in vitro

In vitro
In vitro refers to studies in experimental biology that are conducted using components of an organism that have been isolated from their usual biological context in order to permit a more detailed or more convenient analysis than can be done with whole organisms. Colloquially, these experiments...
by either using purified native or recombinant histones. One standard technique of loading the DNA around the histones involves the use of salt dialysis
Dialysis
In medicine, dialysis is a process for removing waste and excess water from the blood, and is primarily used to provide an artificial replacement for lost kidney function in people with renal failure...
. A reaction consisting of the histone octamers and a naked DNA template can be incubated together at a salt concentration of 2 M. By steadily decreasing the salt concentration, the DNA will equilibrate to a position where it is wrapped around the histone octamers, forming nucleosomes. In appropriate conditions, this reconstitution process allows for the nucleosome positioning affinity of a given sequence to be mapped experimentally.

