SUMO protein
Encyclopedia
Small Ubiquitin-like Modifier or SUMO proteins are a family of small proteins that are covalently
attached to and detached from other proteins in cells
to modify their function. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear
-cytosol
ic transport, transcriptional
regulation, apoptosis
, protein stability, response to stress, and progression through the cell cycle
.
SUMO proteins are similar to ubiquitin
, and SUMOylation is directed by an enzymatic cascade analogous to that involved in ubiquitination. In contrast to ubiquitin, SUMO is not used to tag proteins for degradation
. Mature SUMO is produced when the last four amino acids of the C-terminus have been cleaved off to allow formation of an isopeptide bond between the C-terminal glycine residue of SUMO and an acceptor lysine on the target protein.
SUMO family members often have dissimilar names; the SUMO homologue in yeast
, for example, is called SMT3 (suppressor of mif two 3). Several pseudogenes have been reported for this gene.


-cytosol
ic transport, and transcriptional
regulation. Typically, only a small fraction of a given protein is SUMOylated and this modification is rapidly reversed by the action of deSUMOylating enzymes. SUMOylation of target proteins has been shown to cause a number of different outcomes including altered localization and binding partners. The SUMO-1 modification of RanGAP1
(the first identified SUMO substrate) leads to its trafficking from cytosol to nuclear pore complex . The SUMO modification of hNinein leads to its movement from the centrosome
to the nucleus
. In many cases SUMO modification of transcriptional regulators correlates with inhibition of transcription . Refer to the GeneRIFs of the SUMO proteins, e.g. human SUMO-1 , to find out more.
There are 3 confirmed SUMO isoforms in humans; SUMO-1, SUMO-2
and SUMO-3
. SUMO-2/3 show a high degree of similarity to each other and are distinct from SUMO-1. SUMO-4 shows similarity to -2/3 but it is as yet unclear whether it is a pseudogene
or merely restricted in its expression pattern. During mitosis, SUMO-2/3 localize to centromeres and condensed chromosomes, whereas SUMO-1 localizes to the mitotic spindle and spindle midzone, indicating that SUMO paralogs regulate distinct mitotic processes in mammalian cells . One of the major SUMO conjugation products associated with mitotic chromosomes arose from SUMO-2/3 conjugation of topoisomerase II, which is modified exclusively by SUMO-2/3 during mitosis . SUMO-2/3 modifications seem to be involved specifically in the stress response . SUMO-1 and SUMO-2/3 can form mixed chains, however, because SUMO-1 does not contain the internal SUMO consensus sites found in SUMO-2/3, it is thought to terminate these poly-SUMO chains .
Serine 2 of SUMO-1 is phosphorylated, raising the concept of a 'modified modifier' .
in mass
. The exact length and mass varies between SUMO family members and depends on which organism
the protein comes from. Although SUMO has very little sequence identity with ubiquitin at the amino acid level, it has a nearly identical structural fold.
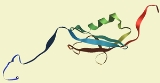
The structure of human SUMO1 is depicted on the right. It shows SUMO1 as a globular protein with both ends of the amino acid chain (shown in red and blue) sticking out of the protein's centre. The spherical core consists of an alpha helix
and a beta sheet
. The diagrams shown are based on an NMR
analysis of the protein in solution.
conjugated to SUMO, x is any amino acid (aa), D or E is an acidic residue. Substrate specificity appears to be derived directly from Ubc9 and the respective substrate
motif. SUMOplot is an online free access software developed to predict the probability for the SUMO consensus sequence (SUMO-CS) to be engaged in SUMO attachment. The SUMOplot score system is based on two criteria: 1) direct amino acid match to the SUMO-CS observed and shown to bind Ubc9, and 2) substitution of the consensus amino acid residues with amino acid residues exhibiting similar hydrophobicity. SUMOplot has been used in the past to predict Ubc9 dependent sites. Seventeen (17) articles have been published so far for the complete list click here. Alternative prediction engines such as SUMOsp are also available .
proteins. Some E3's such as RanBP2 however are neither . Recent evidence has shown that PIAS-gamma is required for the sumoylation of the transcription factor yy1 but it is independent of the zinc-RING finger (identified as the functional domain of the E3 ligases). SUMOylation is reversible and is removed from targets by specific SUMO proteases in an ATP dependent manner. In budding yeast, the Ulp1 SUMO protease is found bound at the nuclear pore, whereas Ulp2 is nucleoplasmic. The distinct subnuclear localisation of deSUMOylating enzymes is conserved in higher eukaryotes
Covalent bond
A covalent bond is a form of chemical bonding that is characterized by the sharing of pairs of electrons between atoms. The stable balance of attractive and repulsive forces between atoms when they share electrons is known as covalent bonding....
attached to and detached from other proteins in cells
Cell (biology)
The cell is the basic structural and functional unit of all known living organisms. It is the smallest unit of life that is classified as a living thing, and is often called the building block of life. The Alberts text discusses how the "cellular building blocks" move to shape developing embryos....
to modify their function. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear
Cell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
-cytosol
Cytosol
The cytosol or intracellular fluid is the liquid found inside cells, that is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrion into compartments....
ic transport, transcriptional
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
regulation, apoptosis
Apoptosis
Apoptosis is the process of programmed cell death that may occur in multicellular organisms. Biochemical events lead to characteristic cell changes and death. These changes include blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, and chromosomal DNA fragmentation...
, protein stability, response to stress, and progression through the cell cycle
Cell cycle
The cell cycle, or cell-division cycle, is the series of events that takes place in a cell leading to its division and duplication . In cells without a nucleus , the cell cycle occurs via a process termed binary fission...
.
SUMO proteins are similar to ubiquitin
Ubiquitin
Ubiquitin is a small regulatory protein that has been found in almost all tissues of eukaryotic organisms. Among other functions, it directs protein recycling.Ubiquitin can be attached to proteins and label them for destruction...
, and SUMOylation is directed by an enzymatic cascade analogous to that involved in ubiquitination. In contrast to ubiquitin, SUMO is not used to tag proteins for degradation
Proteolysis
Proteolysis is the directed degradation of proteins by cellular enzymes called proteases or by intramolecular digestion.-Purposes:Proteolysis is used by the cell for several purposes...
. Mature SUMO is produced when the last four amino acids of the C-terminus have been cleaved off to allow formation of an isopeptide bond between the C-terminal glycine residue of SUMO and an acceptor lysine on the target protein.
SUMO family members often have dissimilar names; the SUMO homologue in yeast
Yeast
Yeasts are eukaryotic micro-organisms classified in the kingdom Fungi, with 1,500 species currently described estimated to be only 1% of all fungal species. Most reproduce asexually by mitosis, and many do so by an asymmetric division process called budding...
, for example, is called SMT3 (suppressor of mif two 3). Several pseudogenes have been reported for this gene.


Function
SUMO modification of proteins has many functions. Among the most frequent and best studied are protein stability, nuclearCell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
-cytosol
Cytosol
The cytosol or intracellular fluid is the liquid found inside cells, that is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrion into compartments....
ic transport, and transcriptional
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
regulation. Typically, only a small fraction of a given protein is SUMOylated and this modification is rapidly reversed by the action of deSUMOylating enzymes. SUMOylation of target proteins has been shown to cause a number of different outcomes including altered localization and binding partners. The SUMO-1 modification of RanGAP1
RANGAP1
Ran GTPase-activating protein 1 is an enzyme that in humans is encoded by the RANGAP1 gene.-Interactions:RANGAP1 has been shown to interact with Ran and UBE2I.-Further reading:...
(the first identified SUMO substrate) leads to its trafficking from cytosol to nuclear pore complex . The SUMO modification of hNinein leads to its movement from the centrosome
Centrosome
In cell biology, the centrosome is an organelle that serves as the main microtubule organizing center of the animal cell as well as a regulator of cell-cycle progression. It was discovered by Edouard Van Beneden in 1883...
to the nucleus
Cell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
. In many cases SUMO modification of transcriptional regulators correlates with inhibition of transcription . Refer to the GeneRIFs of the SUMO proteins, e.g. human SUMO-1 , to find out more.
There are 3 confirmed SUMO isoforms in humans; SUMO-1, SUMO-2
SUMO2
Small ubiquitin-related modifier 2 is a protein that in humans is encoded by the SUMO2 gene.-Further reading:...
and SUMO-3
SUMO3
Small ubiquitin-related modifier 3 is a protein that in humans is encoded by the SUMO3 gene.-Interactions:SUMO3 has been shown to interact with ARNTL and Thymine-DNA glycosylase.-Further reading:...
. SUMO-2/3 show a high degree of similarity to each other and are distinct from SUMO-1. SUMO-4 shows similarity to -2/3 but it is as yet unclear whether it is a pseudogene
Pseudogene
Pseudogenes are dysfunctional relatives of known genes that have lost their protein-coding ability or are otherwise no longer expressed in the cell...
or merely restricted in its expression pattern. During mitosis, SUMO-2/3 localize to centromeres and condensed chromosomes, whereas SUMO-1 localizes to the mitotic spindle and spindle midzone, indicating that SUMO paralogs regulate distinct mitotic processes in mammalian cells . One of the major SUMO conjugation products associated with mitotic chromosomes arose from SUMO-2/3 conjugation of topoisomerase II, which is modified exclusively by SUMO-2/3 during mitosis . SUMO-2/3 modifications seem to be involved specifically in the stress response . SUMO-1 and SUMO-2/3 can form mixed chains, however, because SUMO-1 does not contain the internal SUMO consensus sites found in SUMO-2/3, it is thought to terminate these poly-SUMO chains .
Serine 2 of SUMO-1 is phosphorylated, raising the concept of a 'modified modifier' .
Structure
SUMO proteins are small; most are around 100 amino acids in length and 12 kDaKDA
KDA may refer to:* Karachi Development Authority* Kongsberg Defence & Aerospace* Kotelawala Defence Academy* Kramer Design Associates* Lithium diisopropylamide, KDA is the potassium analogue of lithium diisopropylamideOr kDa may refer to:...
in mass
Mass
Mass can be defined as a quantitive measure of the resistance an object has to change in its velocity.In physics, mass commonly refers to any of the following three properties of matter, which have been shown experimentally to be equivalent:...
. The exact length and mass varies between SUMO family members and depends on which organism
Organism
In biology, an organism is any contiguous living system . In at least some form, all organisms are capable of response to stimuli, reproduction, growth and development, and maintenance of homoeostasis as a stable whole.An organism may either be unicellular or, as in the case of humans, comprise...
the protein comes from. Although SUMO has very little sequence identity with ubiquitin at the amino acid level, it has a nearly identical structural fold.
The structure of human SUMO1 is depicted on the right. It shows SUMO1 as a globular protein with both ends of the amino acid chain (shown in red and blue) sticking out of the protein's centre. The spherical core consists of an alpha helix
Alpha helix
A common motif in the secondary structure of proteins, the alpha helix is a right-handed coiled or spiral conformation, in which every backbone N-H group donates a hydrogen bond to the backbone C=O group of the amino acid four residues earlier...
and a beta sheet
Beta sheet
The β sheet is the second form of regular secondary structure in proteins, only somewhat less common than the alpha helix. Beta sheets consist of beta strands connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet...
. The diagrams shown are based on an NMR
NMR
NMR may refer to:Applications of Nuclear Magnetic Resonance:* Nuclear magnetic resonance* NMR spectroscopy* Solid-state nuclear magnetic resonance* Protein nuclear magnetic resonance spectroscopy* Proton NMR* Carbon-13 NMR...
analysis of the protein in solution.
Prediction of SUMO attachment
Most SUMO-modified proteins contain the tetrapeptide consensus motif Ψ-K-x-D/E where Ψ is a hydrophobic residue, K is the lysineLysine
Lysine is an α-amino acid with the chemical formula HO2CCH4NH2. It is an essential amino acid, which means that the human body cannot synthesize it. Its codons are AAA and AAG....
conjugated to SUMO, x is any amino acid (aa), D or E is an acidic residue. Substrate specificity appears to be derived directly from Ubc9 and the respective substrate
Substrate (biochemistry)
In biochemistry, a substrate is a molecule upon which an enzyme acts. Enzymes catalyze chemical reactions involving the substrate. In the case of a single substrate, the substrate binds with the enzyme active site, and an enzyme-substrate complex is formed. The substrate is transformed into one or...
motif. SUMOplot is an online free access software developed to predict the probability for the SUMO consensus sequence (SUMO-CS) to be engaged in SUMO attachment. The SUMOplot score system is based on two criteria: 1) direct amino acid match to the SUMO-CS observed and shown to bind Ubc9, and 2) substitution of the consensus amino acid residues with amino acid residues exhibiting similar hydrophobicity. SUMOplot has been used in the past to predict Ubc9 dependent sites. Seventeen (17) articles have been published so far for the complete list click here. Alternative prediction engines such as SUMOsp are also available .
SUMO Attachment
SUMO attachment to its target is similar to that of ubiquitin (as it is for the other ubiquitin-like proteins such as NEDD 8). A C-terminal peptide is cleaved from SUMO by a protease (in human these are the SENP proteases or Ulp1 in yeast) using ATP to reveal a di-glycine motif. SUMO then becomes bound to an E1 enzyme (SUMO Activating Enzyme (SAE)) which is a heterodimer. It is then passed to an E2 which is a conjugating enzyme (Ubc9). Finally, one of a small number of E3 ligating proteins attaches it to the protein. In yeast, there are four SUMO E3 proteins, Cst9, Mms21, Siz1 and Siz2. While in ubiquitination an E3 is essential to add ubiquitin to its target, evidence suggests that the E2 is sufficient in Sumoylation as long as the consensus sequence is present. It is thought that the E3 ligase promotes the efficiency of sumoylation and in some cases has been shown to direct SUMO conjugation onto non-consensus motifs. E3 enzymes can be largely classed into PIAS proteins, such as Mms21 (a member of the Smc5/6 complex) and Pias-gamma and HECTHECT domain
In molecular biology, the HECT domain is a protein domain found in ubiquitin-protein ligases. The name HECT comes from Homologous to the E6-AP Carboxyl Terminus'. Proteins containing this domain at the C terminus include ubiquitin-protein ligase, which regulates ubiquitination of...
proteins. Some E3's such as RanBP2 however are neither . Recent evidence has shown that PIAS-gamma is required for the sumoylation of the transcription factor yy1 but it is independent of the zinc-RING finger (identified as the functional domain of the E3 ligases). SUMOylation is reversible and is removed from targets by specific SUMO proteases in an ATP dependent manner. In budding yeast, the Ulp1 SUMO protease is found bound at the nuclear pore, whereas Ulp2 is nucleoplasmic. The distinct subnuclear localisation of deSUMOylating enzymes is conserved in higher eukaryotes
External links
- LifeSensors' SUMO-based Protein and Peptide Expression Systems
- Boston Biochem overview of SUMO reagents and the SUMOylation Cycle
- SUMO1 homology group from HomoloGene
- human SUMO proteins on ExPASy: SUMO1 SUMO2 SUMO3 SUMO4
- UniProt entry for rat Sumo1
Research laboratories
- Ron Hay's lab
- Mary Beth Mudgett's lab (plants & bacterial infection)
- Peter O'Hare's lab (Herpes virus)
- Mary Dasso's section on cell cylce control
- Michael Matunis' lab
- Mary Ann Handel's lab (meiosis, spermatogenesis)
- Nam-Hai Chua's lab (plants, protein modification)
- Frauke Melchior's personal page
- Stefan Jentsch's lab
- Chris Lima's lab
- Bones lab (plant immunology) has a summary page on sumoylation
- SUMOplot Analysis Program — predicts and scores sumoylation sites in your protein (by Abgent)

