
Single linkage clustering
Encyclopedia
In cluster analysis, single linkage, nearest neighbour or shortest distance is a method of calculating distances between clusters in hierarchical clustering
. In single linkage, the distance between two clusters is computed as the distance between the two closest elements in the two clusters.
Mathematically, the linkage function – the distance D(X,Y) between clusters X and Y – is described by the expression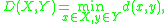
where X and Y are any two sets of elements considered as clusters, and d(x,y) denotes the distance between the two elements x and y.
A drawback of this method is the so-called chaining phenomenon: clusters may be forced together due to single elements being close to each other, even though many of the elements in each cluster may be very distant to each other.
scheme that erases rows and columns in a proximity matrix as old clusters are merged into new ones. The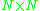 proximity matrix D contains all distances d(i,j). The clusterings are assigned sequence numbers 0,1,......, (n − 1) and L(k) is the level of the kth clustering. A cluster with sequence number m is denoted (m) and the proximity between clusters (r) and (s) is denoted d[(r),(s)].
proximity matrix D contains all distances d(i,j). The clusterings are assigned sequence numbers 0,1,......, (n − 1) and L(k) is the level of the kth clustering. A cluster with sequence number m is denoted (m) and the proximity between clusters (r) and (s) is denoted d[(r),(s)].
The algorithm is composed of the following steps:
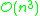 . In 1973, R. Sibson proposed an optimally efficient algorithm of only complexity
. In 1973, R. Sibson proposed an optimally efficient algorithm of only complexity 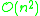 known as SLINK.
known as SLINK.
for minimum spanning tree
s. However, in single linkage clustering, the order in which clusters are formed is important, while for minimum spanning trees what matters is the set of pairs of points that form distances chosen by the algorithm.
Alternative linkage schemes include complete linkage and Average linkage clustering
- implementing a different linkage in the naive algorithm is simply a matter of using a different formula to calculate inter-cluster distances in the initial computation of the proximity matrix and in step 4 of the above algorithm. An optimally efficient algorithm is however not available for arbitrary linkages. The formula that should be adjusted has been highlighted using bold text.
Hierarchical clustering
In statistics, hierarchical clustering is a method of cluster analysis which seeks to build a hierarchy of clusters. Strategies for hierarchical clustering generally fall into two types:...
. In single linkage, the distance between two clusters is computed as the distance between the two closest elements in the two clusters.
Mathematically, the linkage function – the distance D(X,Y) between clusters X and Y – is described by the expression
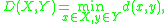
where X and Y are any two sets of elements considered as clusters, and d(x,y) denotes the distance between the two elements x and y.
A drawback of this method is the so-called chaining phenomenon: clusters may be forced together due to single elements being close to each other, even though many of the elements in each cluster may be very distant to each other.
Naive Algorithm
The following algorithm is an agglomerativeHierarchical clustering
In statistics, hierarchical clustering is a method of cluster analysis which seeks to build a hierarchy of clusters. Strategies for hierarchical clustering generally fall into two types:...
scheme that erases rows and columns in a proximity matrix as old clusters are merged into new ones. The
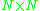 proximity matrix D contains all distances d(i,j). The clusterings are assigned sequence numbers 0,1,......, (n − 1) and L(k) is the level of the kth clustering. A cluster with sequence number m is denoted (m) and the proximity between clusters (r) and (s) is denoted d[(r),(s)].
proximity matrix D contains all distances d(i,j). The clusterings are assigned sequence numbers 0,1,......, (n − 1) and L(k) is the level of the kth clustering. A cluster with sequence number m is denoted (m) and the proximity between clusters (r) and (s) is denoted d[(r),(s)].The algorithm is composed of the following steps:
- Begin with the disjoint clustering having level L(0) = 0 and sequence number m = 0.
- Find the most similar pair of clusters in the current clustering, say pair (r), (s), according to d[(r),(s)] = min d[(i),(j)] where the minimum is over all pairs of clusters in the current clustering.
- Increment the sequence number: m = m + 1. Merge clusters (r) and (s) into a single cluster to form the next clustering m. Set the level of this clustering to L(m) = d[(r),(s)]
- Update the proximity matrix, D, by deleting the rows and columns corresponding to clusters (r) and (s) and adding a row and column corresponding to the newly formed cluster. The proximity between the new cluster, denoted (r,s) and old cluster (k) is defined as d[(k), (r,s)] = min d[(k),(r)], d[(k),(s)].
- If all objects are in one cluster, stop. Else, go to step 2.
Optimally efficient algorithm
The algorithm explained above is easy to understand but of complexity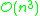 . In 1973, R. Sibson proposed an optimally efficient algorithm of only complexity
. In 1973, R. Sibson proposed an optimally efficient algorithm of only complexity 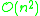 known as SLINK.
known as SLINK.Other linkages
This is essentially the same as Kruskal's algorithmKruskal's algorithm
Kruskal's algorithm is an algorithm in graph theory that finds a minimum spanning tree for a connected weighted graph. This means it finds a subset of the edges that forms a tree that includes every vertex, where the total weight of all the edges in the tree is minimized...
for minimum spanning tree
Minimum spanning tree
Given a connected, undirected graph, a spanning tree of that graph is a subgraph that is a tree and connects all the vertices together. A single graph can have many different spanning trees...
s. However, in single linkage clustering, the order in which clusters are formed is important, while for minimum spanning trees what matters is the set of pairs of points that form distances chosen by the algorithm.
Alternative linkage schemes include complete linkage and Average linkage clustering
UPGMA
UPGMA is a simple agglomerative or hierarchical clustering method used in bioinformatics for the creation of phenetic trees...
- implementing a different linkage in the naive algorithm is simply a matter of using a different formula to calculate inter-cluster distances in the initial computation of the proximity matrix and in step 4 of the above algorithm. An optimally efficient algorithm is however not available for arbitrary linkages. The formula that should be adjusted has been highlighted using bold text.

