.gif)
Splicing (genetics)
Encyclopedia
In molecular biology
and genetics
, splicing is a modification of an RNA
after transcription
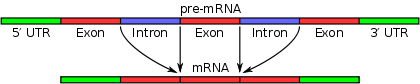
, in which introns are removed and exons are joined. This is needed for the typical eukaryotic messenger RNA
before it can be used to produce a correct protein through translation. For many eukaryotic introns, splicing is done in a series of reactions which are catalyzed
by the spliceosome
, a complex of small nuclear ribonucleoproteins (snRNP
s), but there are also self-splicing introns.
s (C and U), or polypyrimidine tract. Upstream from the polypyrimidine tract is the branch point, which includes an adenine nucleotide. Point mutations in the underlying DNA or errors during transcription can activate a "cryptic splice site" in part of the transcript that usually is not spliced. This results in a mature messenger RNA
with a missing section of an exon. In this way a point mutation
, which usually only affects a single amino acid, can manifest as a deletion
in the final protein.
which is a large RNA-protein complex composed of five small nuclear ribonucleoproteins (snRNP
s, pronounced 'snurps' ). The RNA components of snRNPs interact with the intron and may be involved in catalysis. Two types of spliceosomes have been identified (the major and minor) which contain different snRNP
s.
, performing the functions of the spliceosome by RNA alone. There are three kinds of self-splicing introns, Group I
, Group II
and Group III
. Group I and II introns perform splicing similar to the spliceosome without requiring any protein. This similarity suggests that Group I and II introns may be evolutionarily related to the spliceosome. Self-splicing may also be very ancient, and may have existed in an RNA world present before protein. Although the two splicing mechanisms described below do not require any proteins to occur, 5 additional RNA molecules and over 50 proteins are used and hydrolyzes many ATP molecules. The splicing mechanisms use ATP in order to accurately splice mRNA's. If the cell were to not use any ATP's, the process would be highly inaccurate and many mistakes would occur.
Two transesterifications characterize the mechanism in which group I introns are spliced:
The mechanism in which group II introns are spliced (two transesterification reaction like group I introns) is as follows:
s cleave the RNA and ligases join the exons together.
s or domain
s of life, however, the extent and types of splicing can be very different between the major divisions. Eukaryote
s splice many protein-coding messenger RNAs and some non-coding RNA
s. Prokaryote
s, on the other hand, splice rarely and mostly non-coding RNAs. Another important difference between these two groups of organisms is that prokaryotes completely lack the spliceosomal pathway.
Because spliceosomal introns are not conserved in all species, there is debate concerning when spliceosomal splicing evolved. Two models have been proposed: the intron late and intron early models (see intron evolution).
reactions that occur between RNA nucleotides. tRNA splicing, however, is an exception and does not occur by transesterification.
Spliceosomal and self-splicing transesterification reactions occur via two sequential transesterification reactions. First, the 2'OH of a specific branch-point nucleotide within the intron that is defined during spliceosome assembly performs a nucleophilic attack on the first nucleotide of the intron at the 5' splice site forming the lariat intermediate. Second, the 3'OH of the released 5' exon then performs a nucleophilic attack at the last nucleotide of the intron at the 3' splice site thus joining the exons and releasing the intron lariat.
. Alternative splicing can occur in many ways. Exons can be extended or skipped, or introns can be retained.
s or Peptide nucleic acids to snRNP binding sites, to the branchpoint nucleotide that closes the lariat, or to splice-regulatory element binding sites.
Many splicing errors are safeguarded by a cellular quality control mechanism termed Nonsense-mediated mRNA decay [NMD].
s instead of introns, are removed. The remaining parts, called exteins instead of exons, are fused together.
Protein splicing has been observed in a wide range of organisms, including bacteria, archaea
, plants, yeast and humans.
Molecular biology
Molecular biology is the branch of biology that deals with the molecular basis of biological activity. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry...
and genetics
Genetics
Genetics , a discipline of biology, is the science of genes, heredity, and variation in living organisms....
, splicing is a modification of an RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
after transcription
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
, in which introns are removed and exons are joined. This is needed for the typical eukaryotic messenger RNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
before it can be used to produce a correct protein through translation. For many eukaryotic introns, splicing is done in a series of reactions which are catalyzed
Catalysis
Catalysis is the change in rate of a chemical reaction due to the participation of a substance called a catalyst. Unlike other reagents that participate in the chemical reaction, a catalyst is not consumed by the reaction itself. A catalyst may participate in multiple chemical transformations....
by the spliceosome
Spliceosome
A spliceosome is a complex of snRNA and protein subunits that removes introns from a transcribed pre-mRNA segment. This process is generally referred to as splicing.-Composition:...
, a complex of small nuclear ribonucleoproteins (snRNP
SnRNP
snRNPs , or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs...
s), but there are also self-splicing introns.

Splicing pathways
Several methods of RNA splicing occur in nature; the type of splicing depends on the structure of the spliced intron and the catalysts required for splicing to occur.Spliceosomal introns
Spliceosomal introns often reside in between eukaryotic protein-coding genes. Within the intron, a 3' splice site, 5' splice site, and branch site are required for splicing. The 5' splice site or splice donor site includes an almost invariant sequence GU at the 5' end of the intron, within a larger, less highly conserved consensus region. The 3' splice site or splice acceptor site terminates the intron with an almost invariant AG sequence. Upstream (5'-ward) from the AG there is a region high in pyrimidinePyrimidine
Pyrimidine is a heterocyclic aromatic organic compound similar to benzene and pyridine, containing two nitrogen atoms at positions 1 and 3 of the six-member ring...
s (C and U), or polypyrimidine tract. Upstream from the polypyrimidine tract is the branch point, which includes an adenine nucleotide. Point mutations in the underlying DNA or errors during transcription can activate a "cryptic splice site" in part of the transcript that usually is not spliced. This results in a mature messenger RNA
Mature messenger RNA
Mature messenger RNA, often abbreviated as mature mRNA is a eukaryotic RNA transcript that has been spliced and processed and is ready for translation in the course of protein synthesis...
with a missing section of an exon. In this way a point mutation
Point mutation
A point mutation, or single base substitution, is a type of mutation that causes the replacement of a single base nucleotide with another nucleotide of the genetic material, DNA or RNA. Often the term point mutation also includes insertions or deletions of a single base pair...
, which usually only affects a single amino acid, can manifest as a deletion
Indel
Indel is a molecular biology term that has different definitions in different fields:*In evolutionary studies, indel is used to mean an insertion or a deletion and indels simply refers to the mutation class that includes both insertions, deletions, and the combination thereof, including insertion...
in the final protein.
Spliceosome formation and activity
Splicing is catalyzed by the spliceosomeSpliceosome
A spliceosome is a complex of snRNA and protein subunits that removes introns from a transcribed pre-mRNA segment. This process is generally referred to as splicing.-Composition:...
which is a large RNA-protein complex composed of five small nuclear ribonucleoproteins (snRNP
SnRNP
snRNPs , or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs...
s, pronounced 'snurps' ). The RNA components of snRNPs interact with the intron and may be involved in catalysis. Two types of spliceosomes have been identified (the major and minor) which contain different snRNP
SnRNP
snRNPs , or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs...
s.
- Major
- The major spliceosome splices introns containing GU at the 5' splice site and AG at the 3' splice site. It is composed of the U1U1 spliceosomal RNAU1 spliceosomal RNA is the small nuclear RNA component of U1 snRNP , an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs...
, U2U2 spliceosomal RNAU2 spliceosomal RNA is a small nuclear RNA component of the spliceosome . Complementary binding between U2 snRNA and the branchpoint sequence of the intron results in the bulging out of an unpaired adenosine, on the BPS, which initiates a nucleophilic attack at the intronic 5' splice...
, U4U4 spliceosomal RNAThe U4 small nuclear Ribo-Nucleic Acid is a non-coding RNA component of the major or U2-dependent spliceosome – a eukaryotic molecular machine involved in the splicing of pre-messenger RNA...
, U5U5 spliceosomal RNAU5 RNA is a non-coding RNA that is a component of both types of known spliceosome. The precise function of this molecule is unknown, though it is known that the 5' loop is required for splice site selection and p220 binding, and that both the 3' stem-loop and the Sm site are important for Sm...
, and U6U6 spliceosomal RNAU6 snRNA is the non-coding small nuclear RNA component of U6 snRNP , an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs...
snRNPSnRNPsnRNPs , or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs...
s and is active in the nucleus. In addition, a number of proteins including U2AFU2AF2Splicing factor U2AF 65 kDa subunit is a protein that in humans is encoded by the U2AF2 gene.-Interactions:U2AF2 has been shown to interact with WT1, PUF60, SFRS2IP, SRPK2, SF1, U2 small nuclear RNA auxiliary factor 1 and SFRS11.-References:...
and SF1SF1 (gene)Splicing factor 1 also known as zinc finger protein 162 is a protein that in humans is encoded by the SF1 gene.Splicing factor SF1 is involved in the ATP-dependent formation of the spliceosome complex.-Interactions:...
are required for the assembly of the spliceosome.- E Complex-U1 binds to the GU sequence at the 5' splice site, along with accessory proteins/enzymes ASF/SF2, U2AF (binds at the Py-AG site), SF1/BBP (BBP=Branch Binding Protein);
- A Complex-U2 binds to the branch site and ATP is hydrolyzed;
- B1 Complex-U5/U4/U6 trimer binds, and the U5 binds exons at the 5' site, with U6 binding to U2;
- B2 Complex-U1 is released, U5 shifts from exon to intron and the U6 binds at the 5' splice site;
- C1 Complex-U4 is released, U6/U2 catalyzes transesterification, that make 5'end of introns ligate to the A on intron and form a lariat ,U5 binds exon at 3' splice site, and the 5' site is cleaved, resulting in the formation of the lariat;
- C2 Complex-U2/U5/U6 remain bound to the lariat, and the 3' site is cleaved and exons are ligated using ATP hydrolysis. The spliced RNA is released and the lariat debranches.
- This type of splicing is termed canonical splicing or termed the lariat pathway, which accounts for more than 99% of splicing. By contrast, when the intronic flanking sequences do not follow the GU-AG rule, noncanonical splicing is said to occur (see "minor spliceosome" below).
- Minor
- The minor spliceosomeMinor spliceosomeThe minor spliceosome is a ribonucleoprotein complex that catalyses the removal of an atypical class of spliceosomal introns from eukaryotic messenger RNAs in plant, insects, vertebrates and some fungi . This process is called noncanonical splicing, as opposed to U2-dependent canonical splicing...
is very similar to the major spliceosome, however it splices out rare introns with different splice site sequences. While the minor and major spliceosomes contain the same U5 snRNPSnRNPsnRNPs , or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs...
, the minor spliceosome has different, but functionally analogous snRNPs for U1, U2, U4, and U6, which are respectively called U11U11 spliceosomal RNAThe U11 spliceosomal RNA is a non-coding RNA that together with U4atac/U6atac, U5, and U12 snRNAs and associated proteins, forms a spliceosome that cleaves a divergent class of low-abundance pre-mRNA introns.-References:* [1] * [2]...
, U12U12 minor spliceosomal RNAU12 minor spliceosomal RNA is formed from U12 small nuclear , together with U4atac/U6atac, U5, and U11 snRNAs and associated proteins, forms a spliceosome that cleaves a divergent class of low-abundance pre-mRNA introns. Although the U12 sequence is very divergent from that of U2, the two are...
, U4atac, and U6atac. Like the major spliceosome, it is only found in the nucleus.- Trans-splicing
- Trans-splicingTrans-splicingTrans-splicing is a special form of RNA processing in eukaryotes where exons from two different primary RNA transcripts are joined end to end and ligated....
is a form of splicing that joins two exons that are not within the same RNA transcript.
Self-splicing
Self-splicing occurs for rare introns that form a ribozymeRibozyme
A ribozyme is an RNA molecule with a well defined tertiary structure that enables it to catalyze a chemical reaction. Ribozyme means ribonucleic acid enzyme. It may also be called an RNA enzyme or catalytic RNA. Many natural ribozymes catalyze either the hydrolysis of one of their own...
, performing the functions of the spliceosome by RNA alone. There are three kinds of self-splicing introns, Group I
Group I catalytic intron
Group I introns are large self-splicing ribozymes. They catalyze their own excision from mRNA, tRNA and rRNA precursors in a wide range of organisms. The core secondary structure consists of nine paired regions...
, Group II
Group II intron
Group II introns are a large class of self-catalytic ribozymes as well as mobile genetic element found within the genes of all three domains of life. Ribozyme activity can occur under high-salt conditions in vitro. However, assistance from proteins is required for in vivo splicing...
and Group III
Group III intron
Group III intron is a class of introns found in mRNA genes of chloroplasts in euglenoid protists. They have a conventional group II-type dVI with a bulged adenosine, a streamlined dI, no dII-dV, and a relaxed splice site consensus. Splicing is by two transesterification reactions with a dVI bulged...
. Group I and II introns perform splicing similar to the spliceosome without requiring any protein. This similarity suggests that Group I and II introns may be evolutionarily related to the spliceosome. Self-splicing may also be very ancient, and may have existed in an RNA world present before protein. Although the two splicing mechanisms described below do not require any proteins to occur, 5 additional RNA molecules and over 50 proteins are used and hydrolyzes many ATP molecules. The splicing mechanisms use ATP in order to accurately splice mRNA's. If the cell were to not use any ATP's, the process would be highly inaccurate and many mistakes would occur.
Two transesterifications characterize the mechanism in which group I introns are spliced:
- 3'OH of a free guanine nucleoside (or one located in the intron) or a nucleotide cofactor (GMP, GDP, GTP) attacks phosphate at the 5' splice site.
- 3'OH of the 5'exon becomes a nucleophile and the second transesterification results in the joining of the two exons.
The mechanism in which group II introns are spliced (two transesterification reaction like group I introns) is as follows:
- The 2'OH of a specific adenosine in the intron attacks the 5' splice site, thereby forming the lariat
- The 3'OH of the 5' exon triggers the second transesterification at the 3' splice site thereby joining the exons together.
tRNA splicing
tRNA (also tRNA-like) splicing is another rare form of splicing that usually occurs in tRNA. The splicing reaction involves a different biochemistry than the spliceomsomal and self-splicing pathways. RibonucleaseRibonuclease
Ribonuclease is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 and 3.1 classes of enzymes.-Function:All organisms studied contain...
s cleave the RNA and ligases join the exons together.
Evolution
Splicing occurs in all the kingdomKingdom (biology)
In biology, kingdom is a taxonomic rank, which is either the highest rank or in the more recent three-domain system, the rank below domain. Kingdoms are divided into smaller groups called phyla or divisions in botany...
s or domain
Domain (biology)
In biological taxonomy, a domain is the highest taxonomic rank of organisms, higher than a kingdom. According to the three-domain system of Carl Woese, introduced in 1990, the Tree of Life consists of three domains: Archaea, Bacteria and Eukarya...
s of life, however, the extent and types of splicing can be very different between the major divisions. Eukaryote
Eukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
s splice many protein-coding messenger RNAs and some non-coding RNA
Non-coding RNA
A non-coding RNA is a functional RNA molecule that is not translated into a protein. Less-frequently used synonyms are non-protein-coding RNA , non-messenger RNA and functional RNA . The term small RNA is often used for short bacterial ncRNAs...
s. Prokaryote
Prokaryote
The prokaryotes are a group of organisms that lack a cell nucleus , or any other membrane-bound organelles. The organisms that have a cell nucleus are called eukaryotes. Most prokaryotes are unicellular, but a few such as myxobacteria have multicellular stages in their life cycles...
s, on the other hand, splice rarely and mostly non-coding RNAs. Another important difference between these two groups of organisms is that prokaryotes completely lack the spliceosomal pathway.
Because spliceosomal introns are not conserved in all species, there is debate concerning when spliceosomal splicing evolved. Two models have been proposed: the intron late and intron early models (see intron evolution).
| Eukaryotes | Prokaryotes | |
| Spliceosomal | + | - |
| Self-splicing | + | + |
| tRNA | + | + |
Biochemical mechanism
Spliceosomal splicing and self-splicing involves a two-step biochemical process. Both steps involve transesterificationTransesterification
In organic chemistry, transesterification is the process of exchanging the organic group R″ of an ester with the organic group R′ of an alcohol. These reactions are often catalyzed by the addition of an acid or base catalyst...
reactions that occur between RNA nucleotides. tRNA splicing, however, is an exception and does not occur by transesterification.
Spliceosomal and self-splicing transesterification reactions occur via two sequential transesterification reactions. First, the 2'OH of a specific branch-point nucleotide within the intron that is defined during spliceosome assembly performs a nucleophilic attack on the first nucleotide of the intron at the 5' splice site forming the lariat intermediate. Second, the 3'OH of the released 5' exon then performs a nucleophilic attack at the last nucleotide of the intron at the 3' splice site thus joining the exons and releasing the intron lariat.
Alternative splicing
In many cases, the splicing process can create a range of unique proteins by varying the exon composition of the same messenger RNA. This phenomenon is then called alternative splicingAlternative splicing
Alternative splicing is a process by which the exons of the RNA produced by transcription of a gene are reconnected in multiple ways during RNA splicing...
. Alternative splicing can occur in many ways. Exons can be extended or skipped, or introns can be retained.
Experimental manipulation of splicing
Splicing events can be experimentally altered by binding steric-blocking antisense oligos such as MorpholinoMorpholino
In molecular biology, a Morpholino is a molecule in a particular structural family that is used to modify gene expression. Morpholino oligomers are an antisense technology used to block access of other molecules to specific sequences within nucleic acid...
s or Peptide nucleic acids to snRNP binding sites, to the branchpoint nucleotide that closes the lariat, or to splice-regulatory element binding sites.
Splicing errors
Common errors:- Mutation of a splice site resulting in loss of function of that site. Results in exposure of a premature stop codonStop codonIn the genetic code, a stop codon is a nucleotide triplet within messenger RNA that signals a termination of translation. Proteins are based on polypeptides, which are unique sequences of amino acids. Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide...
, loss of an exon, or inclusion of an intron. - Mutation of a splice site reducing specificity. May result in variation in the splice location, causing insertion or deletion of amino acids, or most likely, a disruption of the reading frameReading frameIn biology, a reading frame is a way of breaking a sequence of nucleotides in DNA or RNA into three letter codons which can be translated in amino acids. There are 3 possible reading frames in an mRNA strand: each reading frame corresponding to starting at a different alignment...
. - Displacement of a splice site, leading to inclusion or exclusion of more RNA than expected, resulting in longer or shorter exons.
Many splicing errors are safeguarded by a cellular quality control mechanism termed Nonsense-mediated mRNA decay [NMD].
Protein splicing
In addition to RNA, proteins can undergo splicing. Although the biomolecular mechanisms are different, the principle is the same: parts of the protein, called inteinIntein
An intein is a segment of a protein that is able to excise itself and rejoin the remaining portions with a peptide bond. Inteins have also been called "protein introns"....
s instead of introns, are removed. The remaining parts, called exteins instead of exons, are fused together.
Protein splicing has been observed in a wide range of organisms, including bacteria, archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
, plants, yeast and humans.
See also
- cDNA
- ExonExonAn exon is a nucleic acid sequence that is represented in the mature form of an RNA molecule either after portions of a precursor RNA have been removed by cis-splicing or when two or more precursor RNA molecules have been ligated by trans-splicing. The mature RNA molecule can be a messenger RNA...
- IntronIntronAn intron is any nucleotide sequence within a gene that is removed by RNA splicing to generate the final mature RNA product of a gene. The term intron refers to both the DNA sequence within a gene, and the corresponding sequence in RNA transcripts. Sequences that are joined together in the final...
- Primary transcriptPrimary transcriptA primary transcript is an RNA molecule that has not yet undergone any modification after its synthesis. For example, a precursor messenger RNA is a primary transcript that becomes a messenger RNA after processing, and a primary microRNA precursor becomes a microRNA after processing....
- SpliceosomeSpliceosomeA spliceosome is a complex of snRNA and protein subunits that removes introns from a transcribed pre-mRNA segment. This process is generally referred to as splicing.-Composition:...
- Minor spliceosomeMinor spliceosomeThe minor spliceosome is a ribonucleoprotein complex that catalyses the removal of an atypical class of spliceosomal introns from eukaryotic messenger RNAs in plant, insects, vertebrates and some fungi . This process is called noncanonical splicing, as opposed to U2-dependent canonical splicing...
- Exon Junction ComplexExon junction complexThe exon junction complex has major influences on translation, surveillance and localization of the spliced mRNA. It is first deposited onto mRNA during splicing and is then transported into the cytoplasm. There it plays a major role in post-transcriptional regulation of mRNA. It is believed that...

