
Wobble base pair
Encyclopedia
Molecular biology
Molecular biology is the branch of biology that deals with the molecular basis of biological activity. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry...
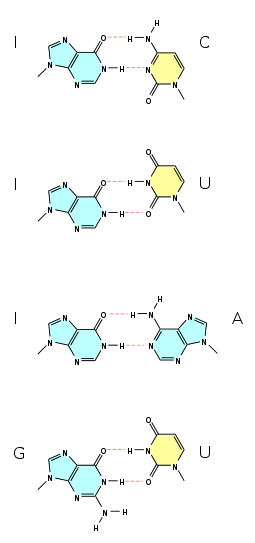
, a wobble base pair is a non-Watson-Crick base pairing between two nucleotides in RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
molecules. The four main wobble base pairs are guanine
Guanine
Guanine is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine . In DNA, guanine is paired with cytosine. With the formula C5H5N5O, guanine is a derivative of purine, consisting of a fused pyrimidine-imidazole ring system with...
-uracil
Uracil
Uracil is one of the four nucleobases in the nucleic acid of RNA that are represented by the letters A, G, C and U. The others are adenine, cytosine, and guanine. In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine.Uracil is a common and...
, inosine
Inosine
Inosine is a nucleoside that is formed when hypoxanthine is attached to a ribose ring via a β-N9-glycosidic bond....
-uracil
Uracil
Uracil is one of the four nucleobases in the nucleic acid of RNA that are represented by the letters A, G, C and U. The others are adenine, cytosine, and guanine. In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine.Uracil is a common and...
, inosine
Inosine
Inosine is a nucleoside that is formed when hypoxanthine is attached to a ribose ring via a β-N9-glycosidic bond....
-adenine
Adenine
Adenine is a nucleobase with a variety of roles in biochemistry including cellular respiration, in the form of both the energy-rich adenosine triphosphate and the cofactors nicotinamide adenine dinucleotide and flavin adenine dinucleotide , and protein synthesis, as a chemical component of DNA...
, and inosine
Inosine
Inosine is a nucleoside that is formed when hypoxanthine is attached to a ribose ring via a β-N9-glycosidic bond....
-cytosine
Cytosine
Cytosine is one of the four main bases found in DNA and RNA, along with adenine, guanine, and thymine . It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached . The nucleoside of cytosine is cytidine...
(G-U, I-U, I-A and I-C). The thermodynamic stability of a wobble base pair is comparable to that of a Watson-Crick base pair. Wobble base pairs are fundamental in RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
secondary structure
Secondary structure
In biochemistry and structural biology, secondary structure is the general three-dimensional form of local segments of biopolymers such as proteins and nucleic acids...
and are critical for the proper translation of the genetic code
Genetic code
The genetic code is the set of rules by which information encoded in genetic material is translated into proteins by living cells....
.
tRNA wobble
In the genetic codeGenetic code
The genetic code is the set of rules by which information encoded in genetic material is translated into proteins by living cells....
, there are
 = 64 possible codons (tri-nucleotide
= 64 possible codons (tri-nucleotideNucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
sequences). For translation
Translation (genetics)
In molecular biology and genetics, translation is the third stage of protein biosynthesis . In translation, messenger RNA produced by transcription is decoded by the ribosome to produce a specific amino acid chain, or polypeptide, that will later fold into an active protein...
, each of these codons requires a tRNA
Transfer RNA
Transfer RNA is an adaptor molecule composed of RNA, typically 73 to 93 nucleotides in length, that is used in biology to bridge the three-letter genetic code in messenger RNA with the twenty-letter code of amino acids in proteins. The role of tRNA as an adaptor is best understood by...
molecule with a complementary anticodon. If each tRNA molecule paired with its complementary mRNA codon using canonical Watson-Crick base pairing, then 64 types (species) of tRNA molecule would be required. In the standard genetic code, three of these 64 codons are stop codons, which terminate translation by binding to release factor
Release factor
A release factor is a protein that allows for the termination of translation by recognizing the termination codon or stop codon in a mRNA sequence....
s rather than tRNA molecules, so canonical pairing would require 61 species of tRNA. Since most organisms have fewer than 45 species of tRNA, some tRNA species must pair with more than one codon. In 1966, Francis Crick
Francis Crick
Francis Harry Compton Crick OM FRS was an English molecular biologist, biophysicist, and neuroscientist, and most noted for being one of two co-discoverers of the structure of the DNA molecule in 1953, together with James D. Watson...
proposed the Wobble hypothesis to account for this. He postulated that the 5'
Directionality (molecular biology)
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. The chemical convention of naming carbon atoms in the nucleotide sugar-ring numerically gives rise to a 5′-end and a 3′-end...
base on the anticodon, which binds to the 3'
Directionality (molecular biology)
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. The chemical convention of naming carbon atoms in the nucleotide sugar-ring numerically gives rise to a 5′-end and a 3′-end...
base on the mRNA, was not as spatially confined as the other two bases, and could, thus, have non-standard base pairing.
As an example, yeast
Yeast
Yeasts are eukaryotic micro-organisms classified in the kingdom Fungi, with 1,500 species currently described estimated to be only 1% of all fungal species. Most reproduce asexually by mitosis, and many do so by an asymmetric division process called budding...
tRNAPhe
Phenylalanine
Phenylalanine is an α-amino acid with the formula C6H5CH2CHCOOH. This essential amino acid is classified as nonpolar because of the hydrophobic nature of the benzyl side chain. L-Phenylalanine is an electrically neutral amino acid, one of the twenty common amino acids used to biochemically form...
has the anticodon 5'-GmAA-3' and can recognize the codons 5'-UUC-3' and 5'-UUU-3'. It is, therefore, possible for non-Watson–Crick base pairing to occur at the third codon position, i.e., the 3' nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
of the mRNA codon and the 5' nucleotide of the tRNA anticodon.
tRNA Base pairing schemes
The original wobble pairing rules, as proposed by Crick. Watson-Crick base pairs are shown in bold, wobble base pairs in italic:| tRNA 5' anticodon base | mRNA 3' codon base |
|---|---|
| A | U |
| C | G |
| G | C or U |
| U | A or G |
| I | A or C or U |
Revised pairing rules
| tRNA 5' anticodon base | mRNA 3' codon base |
|---|---|
| G | U,C |
| C | G |
| k2C | A |
| A | U,C,(A),G |
| unmodified U | U,(C),A,G |
| xm5s2U,xm5Um,Um,xm5U | A,(G) |
| xo5U | U,A,G |
| I | A,C,U |

