
Folding@home
Encyclopedia
Folding@home is a distributed computing
project designed to use spare processing power on personal computers to perform simulations of disease-relevant protein folding
and other molecular dynamics
, and to improve on the methods of doing so. Also referred to as FAH or F@h, much of its work attempts to determine how proteins reach their final structure, which is of significant academic interest and has implications to both disease research and nanotechnology
. To a lesser degree Folding@home also tries to predict
that final structure from only the initial amino acid
sequence, which has applications in drug design
. Folding@home is developed and operated by the Pande Laboratory at Stanford University
, under the direction of Vijay Pande. The goal of the project is to "understand protein folding, misfolding, and related diseases".
Folding@home's simulations of protein folding and misfolding enable the scientific community to better understand the development of many diseases, including Alzheimer's disease
, Parkinson's disease
, cancer
, Creutzfeldt–Jakob disease, Huntington's disease
, cystic fibrosis
, sickle-cell anaemia, HIV
, Chagas disease
, influenza
, osteogenesis imperfecta
, autism
, and alpha 1-antitrypsin deficiency
, among others. More fundamentally, understanding the process of protein folding — how proteins assemble themselves into a functional state — is one of the outstanding problems of molecular biology
. The Pande lab has produced ninety-five scientific research papers
as a direct result of the project, and Folding@home has caused paradigm shift
s in protein folding theory.
In January 2010 the Folding@home project successfully simulated protein folding in the 1.5 millisecond
range — a simulation thousands of times longer than ever previously achieved. Folding@home has pioneered the uses of GPUs, multi-core processors, and PlayStation 3
s for distributed computing. It remains one of the world's fastest computing systems, and is more powerful than all distributed computing projects under BOINC combined. Folding@home's distributed simulations remain accurate compared to results from laboratory research, a "grand challenge" in computational biology
. The Pande lab's goal is to refine Folding@home's methods to the level where it will become an essential tool for molecular medical research, and they collaborate with various scientific institutions and laboratories across the world.
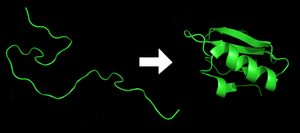 Protein
Protein
s are an essential component to many biological functions, and participate in virtually every process within cells. They often act as enzymes, performing biochemical reactions including cell signaling
, transportation, cellular regulation, and others. As structural elements, some proteins act as a type of skeleton for cells
, and as antibodies other proteins help the immune system
. Before a protein can take on these roles, it must fold into a functional three-dimensional shape based on a particular series of steps
, which often occur spontaneously
. Understanding protein folding is thus critical to understanding what a protein does and how it works. Moreover, when proteins do not fold correctly—that is, when they fold down the wrong pathway and end up misshapen—they can aggregate
and cause serious and in some cases life-threatening diseases. However, once the misfolding process is determined, figuring out its prevention can be the next step, and preventative treatments can be applied during the folding process. While the understanding of protein folding requires a combination of theories and experiments, creating experimentally comparable simulations of protein folding dynamics remains difficult and is considered a "holy grail" of computational biology
. Folding@home's distributed computing methods can address these challenges and allow for unique insights into the complexity of protein folding while strongly agreeing with experimental data.
Since the late 1990s, simulations of protein molecular dynamics have been severely limited by computational power. Straightforward approaches have exceptional difficulty for all but the most elementary of systems, which has prompted the use of simplified native
-centric in silico
molecular models. Not only are these models insufficient for a comprehensive view of protein folding, but the traditional methods of using a small number of very long simulations cannot accurately capture enough detailed information into how a protein misfolds. Moreover, these simplified models often ignore how solvent
s such as water might affect the folding process. In strong agreement with experimental observations, its influence was revealed in 2004 using Folding@home's computational power and explicit solvation simulations. This observation was made possible by Folding@home's statistical assembly of many shorter simulations, which compared to traditional approaches, provide a much more complete description of the protein's energy landscape
, conformation space
, and equilibrium thermodynamics
. While some of these simulations may work themselves into impossible atomic configurations or into the correct native state, others will illustrate how that protein misfolds. By utilizing thermodynamics
, Folding@home uses methods that can find these rare events without having to complete many simulations. This allows for a highly accurate and thorough exploration of protein configurations in a reasonable amount of time, even over very long simulation timescales. For the instrumental development of the software behind these statistical approaches and for attaining quantitative agreement between theory and experiment, in 2010 Folding@home researcher Greg Bowman was awarded the Thomas Kuhn
Paradigm Shift Award from the American Chemical Society
.
The Pande lab and other researchers can use Folding@home to study aspects of folding, misfolding, and related diseases that would never be seen experimentally. For example, most proteins have such intrinsically stable native state
s that it is difficult to experimentally study their folding events, and researchers must resort to chemical denaturation
methods. While the denatured state of proteins is of scientific interest because any residual structure may affect its folding behavior, even with molecular experiments it remains difficult to determine the extent of a local structure that may be present. However, molecular simulations on Folding@home can provide detail into denatured conformational states and insights into the chemical denaturation mechanisms. These and other simulations run on Folding@home are used in conjunction with laboratory experiments. Additionally, as of 2011 the Pande lab is performing studies into how protein folding in their native cells
may be different than in environments such as test tube
s used during experiments.
In addition to the diseases listed below, researchers use Folding@home to study malaria
, Chagas disease
, the prion
s which cause Creutzfeldt–Jakob disease, and details into how virus
es such as HIV
and influenza
function and infect cellular membranes. Results from this research have led to major shifts in the understanding of protein folding and its applications for disease, as well as improved protein folding models. Folding@home is dedicated to producing significant amounts of results towards protein folding, the diseases that result from protein misfolding, and novel computational methods for doing so. The goal of the first five years of the project was to make significant advances in understanding folding, while the current goal is to understand misfolding and related disease, especially Alzheimer's disease.
As a part of Stanford University, a non-profit organization, the Pande lab does not sell the results generated by Folding@home. The large data sets from the project are freely available for other researchers to use upon request, and some can be accessed from the Folding@home website. The Pande lab also releases Folding@home's key software to other researchers, so that the algorithms which benefit Folding@home will also aid other scientific areas. Moreover, in 2011 they released the open-source Copernicus software, so that other researchers can run molecular simulations much more efficiently on clusters or supercomputers. Summaries of all of the scientific findings from Folding@home are posted on the Folding@home website after publication. The full publications are available online or from a local municipal or academic library.
, a form of dementia
which most often affects the elderly, is believed to be caused by specific misfolding and subsequent aggregation
of the small 42-residue
amyloid beta
(Aß) peptide
. The severity of the disease depends not only on the amount of Aß, but also on how it misfolds. Current theory holds that toxic non-plaque
Aß oligomers (aggregates of many monomers) bind to a surface receptor on neuron
s and change the structure of the synapse
, thereby disrupting neuronal communication and causing neuronal cell death which leads to the associated neurodegenerative consequences. Understanding how and why this peptide misfolds could result in key insights into how to cure Alzheimer's Disease, and will also help the Pande lab prepare for similar aggregation studies.
Despite this connection, toxic Aß aggregations remain so complex that it was not previously possible to simulate them in atomic resolution. In 2011, the Pande lab explored how their Abeta studies using Folding@home could be used as a starting point for a new Alzheimer's therapy. Folding@home is currently concentrating on Alzheimer's and continues full-scale simulations of amyloid beta and its oligomerization, which had previously been a technological challenge to simulate. These studies build off of the Pande lab's 2008 published work into new ways to simulate Abeta oligomerization over long timescales. In the same publication, previous all-atom simulations were performed that led to specific experimentally-tested predictions, such as ways in which to stabilize the protein and prevent the toxic oligomer formation. The Pande lab is focusing their research in this area for rational drug design
approaches. Pande described that paper as the "tip of the iceberg" for the Folding@home studies of Alzheimer's, as further results will follow and possibly new therapeutics.
Folding@home is also being used to study Aß fragments of different sizes to determine how various natural enzymes affect the structure and folding of Aß. These fragments are tied to senile plaques, a pathological marker of Alzheimer's disease in patient's brain. When certain enzymes cleave the amyloid precursor protein
, Abeta peptides are produced, while the action of other enzymes can instead produce p3 peptides, much smaller fragments of Aß. Folding@home is simulating one of these smaller peptides in water in an effort to determine how the length of Aß affects its overall structure.
In 2010, several possible drug leads predicted by Folding@home went from the test tube
to testing on living tissue
, and in close cooperation with the Nanomedicine Center for Protein Folding, the drug leads continued to be refined. Additionally, as predicted by FAH's simulations, a stable form of amyloid beta was experimentally verified which the Pande lab believes could be used as a starting point for new Alzheimer's therapy. In 2008, Folding@home produced several small drug candidates to fight Alzheimer's Disease, as they appear to inhibit the toxicity of Abeta.
The Pande lab is also using Folding@home to investigate protein–protein interactions, which occur extensively throughout both benign and disease-related biological activities. Interactions involving the common SH3
protein are also being studied, as it has implications in Alzheimers research. The refinement of these simulations has greatly improved the Pande lab's ability to understand a wide variety of biological interactions.
, an incurable neurodegenerative genetic disorder affecting muscle coordination
and leading to dementia
, is also associated with protein misfolding. Specifically, it is caused by a mutation in the Huntingtin gene, which causes excessively long repetitive chains
of the glutamine
amino acid in the Huntingtin protein, a protein that plays important roles in nerve cells. The likelihood of neuronal cell death is primarily affected by the length of the glutamine chain and the neuron's intracellular exposure to the misfolded Huntingtin protein. The defective protein causes Huntington's by aggregating most often in the striatum
and frontal cortex of patient's brains. The Pande lab is using Folding@home to study these aggregates, as well as predict how they form. How this aggregation occurs has been largely unknown, but in 2009 a paper based off of Folding@home's results and published in the Journal of Molecular Biology
investigated possible mechanisms for the aggregation formation, and the implications into how to prevent it. These studies will be useful for drug design
approaches against the disease, and will serve as a foundation for methods to stop the aggregation formation. Additionally, some of the methods used to study Huntington's are also being used for Alzheimer's research.
In 2010, Folding@home researcher Veena Thomas proposed a novel therapeutic strategy for HD, which may be funded by the NIH
. This strategy could be used to bring the results from Folding@home directly to a therapeutic.
s involve mutations in p53
, a tumor suppressor protein present in every cell which signals for cell death in the event of damage to a cell's DNA
. If p53 becomes mutated, breaks down, or fails to fold properly, it will no longer be able to cause the "stop signal" for cell division. Cells will consequently divide and grow uncontrollably to form tumors. Folding@home is used to study specific properties of p53 in order to understand and predict these mutations, and in 2007 work began to develop inhibitor proteins that deactivate damaged p53. The Pande lab has also developed a novel experimentally-verified method for predicting how mutations affect p53, which has had reasonable success in the identification of deleterious mutations such as those linked to cancer. This supplemented work performed in 2005 that studied how mutations affect the folding of p53, and which mutations are relevant to cancer. While it was the first peer-reviewed publication from a distributed computing project related to cancer, the study also agreed well with experiments and offered insights that were previously unobtainable. Following these results, the Pande lab have expanded their work to other p53-related diseases.
The Pande lab is also performing research into protein chaperones. These are proteins that assist in the folding of other molecules, assembly of oligomeric structures, the prevention of potential damage caused by protein misfolding, and other functions. They are needed for these purposes by rapidly growing cancerous cells. Using Folding@home and working closely with the Protein Folding Center, they plan to find ways to inhibit chaperones involved in cancer. Using Folding@home for a more comprehensive visualization of their functions, the Pande lab and the Protein Folding Center collectively plan to engineer modified chaperonins to inhibit the folding of particular proteins associated with human diseases such as cancer and Alzheimer's. While this approach has been used before, they believe that this project, if successful, could lead to an interesting new drug against cancer or at least make major advances in that area.
Folding@home is also used to study the folding of several other proteins which have mutations tied to cancer, such as the enzyme src Kinase and certain forms of the Engrailed
Homeodomain. These proteins also have a great deal of experimental data for comparison, and serve as a great system for the understanding of folding and misfolding. Additionally, the Pande lab is using Folding@home to understand the dynamics of a small knottin
protein and how it can be used to bind to contrast agents for imaging scan
or drug
s. Finally, some forms of interleukin-2, an important signaling protein for the immune system, have been used as immunotherapy
for cancer. The Pande lab believes that Folding@home's simulations of its dynamics will lead to insights into how to design other therapeutics.
is a degenerative
disorder of the central nervous system
, characterized by shaking, rigidity, slowness of movement, and dementia
. The Pande lab has performed preliminary studies on the properties of alpha-synuclein
, a key natively unfolded protein
. Particular mutations of alpha-synuclein can aggregate to form toxic fibrils, and while the mechanism of this aggregation remains largely unknown, it can lead to the Parkinson's disease and other conditions. The Pande lab is also testing how Folding@home's methods apply to this problem, and in 2005, Pande presented results from FAH at a National Parkinson Foundation
conference.
is a non-curable genetic bone disorder. Those with the disease are unable to successfully make functional connective bone tissue. This is lethal for many but can also induce a higher rate of miscarriages. The disease is caused by mutations in the Type-1 collagen
protein, the most common form of collagen and found abundantly throughout the body. Although some of these mutations of collagen can lead to serious morphological disorders, more benign forms can cause brittle bones and other subtleties. Folding@home has performed simulations of collagen, and has produced a paper on Osteogenesis imperfecta outlining new molecular simulation techniques and revealing new insights into how collagen misfolds. The Pande lab believes these results will be useful for later computational studies of collagen.
is a misfolded peptide
involved in Type II diabetes. While amylin is natively unfolded, it forms an alpha helix
structure upon contact with cellular membranes. Moreover, it can aggregate into large deposits on these membranes, inducing cell death of insulin
-producing cells, which may be relevant to the development of the disease. Around 95% of patients with Type II diabetes exhibit these aggregate deposits. As of 2011, Folding@home is simulating amylin with the goal to understand how this aggregation forms and to design drugs to prevent it.
is a large biological machine that synthesizes proteins from mRNA. It is the target for approximately half of all known antibiotics, which usually kill bacteria by preventing their ribosomes from making new and essential proteins. Folding@home is simulating the ribosome in detail using new state-of-the-art calculation methods. Results from these simulations have significantly helped the Pande lab prepare to study more complex biomedical problems. The Pande lab is also using Folding@home to perform antibiotic drug design calculations.
of small molecules to a protein. Accurate predictions of binding affinities have the potential to significantly lower the development cost of new drugs. Additionally, Folding@home is utilized to find prime binding sites on protein surfaces by simulating interactions between ligand binding sites with different molecules. This has a direct application to computational drug design
. Folding@home is also performing calculations on beta-lactamase
, a protein that plays important roles in drug resistance
. The Pande lab hopes that by understanding its dynamics, they may be able to design drugs to deactivate it.
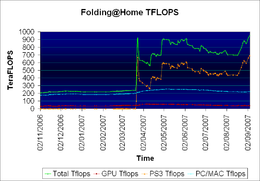 Interest and participation in the project has grown steadily since its launch. As of November 17, 2011, Folding@home has about 439,000 active CPUs, about 37,000 active GPUs, and about 21,000 active PS3s
Interest and participation in the project has grown steadily since its launch. As of November 17, 2011, Folding@home has about 439,000 active CPUs, about 37,000 active GPUs, and about 21,000 active PS3s
, for a total of about 6.7 native petaFLOPS, (9.1 x86 petaFLOPS) more computing power than the combined efforts of all distributed computing projects under BOINC. A large majority of this performance comes from the GPU and PS3 clients. Folding@home achieves strong scaling across its user base; it gains a near-linear speedup for every additional processor. In 2007, Guinness
recognized Folding@home as the most powerful distributed computing cluster in the world. This large and powerful network allows FAH to do work not possible any other way, including through the use of supercomputers, which are typically expensive to operate and often shared.
Folding@home gained popularity early in its history. In March 2002, Google
co-founder Sergey Brin
launched Google Compute as add-on for the Google Toolbar
. Although limited in functionality and scope, it increased Folding@home's participation from 10,000 up to about 30,000 active CPUs. The program ended in October 2005 in favor of the Pande lab's official clients, and is no longer available for the Toolbar. Folding@home also gained participants from Genome@home
, another distributed computing project from the Pande lab and a sister project to Folding@home. The goal of Genome@home was protein design
and its applications, and the project was officially concluded in March 2004. Following its completion, users were asked to donate to Folding@home instead.
On September 16, 2007, the Folding@home project officially attained a sustained performance level higher than one native petaFLOPS
, becoming the first computing system of any kind in the world to do so, although it had erroneously almost reached that level in March of that year. On May 7, 2008, the project attained a sustained performance level higher than two native petaFLOPS, followed by the three and four native petaFLOPS milestones on August 20 and September 28, 2008 respectively. Then on February 18, 2009, Folding@home achieved a performance level of just above five native petaFLOPS
, thereby becoming the first computing system to surpass that performance, just as it was for the other four milestones. Finally, on November 10, 2011, Folding@home crossed the six native petaFLOP barrier with the equivalent of nearly eight x86 petaFLOPS.
Starting in March 2009, Folding@home began reporting performance in both native and x86 FLOPS. While native FLOPS are a measure of the performance from a given hardware, Folding@home also estimates how many FLOPS the calculation would take on the standard x86 architecture, which is commonly used as a performance reference. For instance, certain complex functions can be performed in one native FLOP on a GPU, but take multiple FLOPS on the standard x86 CPU architecture. Despite using conservative conversions, for the GPU and PS3 clients "x86" FLOPS are consistently much greater than the "Native" FLOPS. By reporting in both native and x86 FLOPS, Folding@home attempts to even out these differences.
Users can register their contributions under a team, which register the combined score of all their members. A user can start their own team, or they can join an existing team. They can be used for troubleshooting or recruitment purposes, but can also keep donors motivated. In some cases, a team may have their own community-driven sources of help such as a forum
. In addition, rivalries between teams create friendly competition that benefits the folding community, and members can also have intra-team competitions for top spots. However, regardless of username or team affiliation, all contributions go to the same place and have the same scientific value. Rankings and other statistics for both individuals and teams are posted to the Folding@home website, with third party statistics sites also available.
program
on their personal computer
. The project differs from other distributed computing projects, such as those under BOINC, by offering a variety of clients such as those for multi-core processors, graphics processing unit
s, and PlayStation 3
s, complementing the standard client designed for uniprocessor
systems. While the former clients use significantly more system resources, they also have the capability of completing an overall simulation very quickly, (in a few weeks or months rather than years) which is of major scientific value. Folding@home is the first project to fully utilize GPUs, PS3s, or multi-core processors for distributed computing. As its software is custom-tailored to each hardware architecture
, Folding@home gains the ability to run many different types of calculations, allowing the Pande Group to address questions previously considered impossible to tackle computationally, and make even greater impacts on knowledge of protein misfolding and its related diseases.
Each client is the software with which the user interacts, and manages the other software components behind the scenes. Through the client, the user may pause the folding process, open an event log, check the work progress, or view personal statistics. These clients run continuously in the background
, using otherwise unused processing power. These clients are designed to run FAH's calculations at an extremely low priority, and will back off to allow other computer programs to have more processing power. Although modern computer chips are designed to be able to operate continuously without degrading, if users wish to reduce power consumption or heat production, the maximum percentage of CPU power used can be adjusted if desired. If interrupted by a computer shutdown or other means, the client will resume work at almost the same point at startup. For users with machines with multiple processor units, multiple clients may be installed on one machine, and users may be credited by clients on multiple machines.
For security and scientific integrity reasons, the Pande lab does not publicly release the source code
of the clients. Significant work goes into minimizing security issues in all of Folding@home's software. For example, clients can be downloaded only from the official Folding@home website or its commercial partners. It will upload and download data only from Stanford's Folding@home data servers, (over port
8080, with 80 as an alternative) and will only interact with FAH computer files. Moreover, it does not normally need computer administrative privileges, so from a security standpoint it behaves similar to but is even more secure than a web browser
.
Folding@home's first client was a screensaver
, which would run Folding@home while the computer was not otherwise in use. Later, the Pande lab tested clients on the open source BOINC framework; however, this approach became unworkable and was abandoned in June 2006. Both BOINC and Folding@home clients fell short, for neither client type had enough ability to be compatible with the other. BOINC lacked many features that FAH needed, and FAH lacked features that BOINC needed.
s) at the cost of lower generality. For this reason, high-performance computing is increasingly utilizing the GPU specialized hardware. In the first large-scale test of GPU scientific reliability, the Pande lab found that although GPUs lack built-in memory error detection and correction
, reliable scientific computation can be performed on consumer-grade hardware, as long as sufficient measures are taken (such as Folding@home's built-in error detection) to ensure data integrity.
Despite their potential benefits, scientific computing on GPUs has previously remained inefficient and difficult. The Pande lab has been able to write OpenMM, an open source
molecular dynamics library optimized to take full advantage of the GPU architecture and gain large speed increases over conventional single-CPU implementations. As an abstraction layer
, OpenMM allows molecular dynamics simulations to be efficiently run across a variety of computer architectures and platforms, something previously problematic in scientific software development. GPUs remain the most powerful platform available in terms of FLOPS
; as of September 23, 2011, GPU clients account for 71% of the entire project's FLOP throughput.
The first generation of Folding@home's Windows
GPU client (GPU1) was released to the public on October 2, 2006, and delivered a 20-30X speedup for certain calculations over its CPU-based Gromacs counterparts. It was the first time GPUs had been used for either distributed computing or major molecular dynamics calculations. The Pande lab learned much about the development of GPGPU
software, but citing a need to improve scientific accuracies over DirectX
, it was succeeded by GPU2, the second generation successor of the client on April 10, 2008. Several months later, GPU1 was officially retired on June 6. Compared to GPU1, GPU2 was more scientifically reliable and productive, ran on ATI
and CUDA
-enabled Nvidia
GPUs, and supported more advanced algorithms, larger proteins, and real-time visualization of the protein simulation. Following this, the third generation of Folding@home's GPU client (GPU3) was released on May 25, 2010. While backwards compatible to GPU2, GPU3 is more stable and efficient, has additional scientific capabilities, and uses the Pande lab's OpenMM library on top of an OpenCL
framework. Although it does not natively support the Linux
operating system, it can be run under WINE
for donors with Nvidia graphics cards.
 Folding@home can also take advantage of the computing power of PlayStation 3
Folding@home can also take advantage of the computing power of PlayStation 3
s, to achieve performance previously only possible on supercomputers. Unlike Microsoft's Xbox
, the PS3 is well suited for Folding@home simulations. At the time of its inception and for certain calculations, its main streaming processor
Cell processor delivered a 20x speed increase over PCs, allowing the Pande lab to address problems previously considered impossible to tackle computationally. This high speed and efficiency introduced other opportunities for worthwhile
optimizations, and radically changed the tradeoff between computational efficiency and overall accuracy, allowing for the utilization of more complex molecular models at little extra computational cost. These capabilities allow for greater insights into disease research.
The PS3 client was originally a standalone application, but since September 18, 2008 is a channel of Life with PlayStation
, developed in a collaborative effort between Sony
and the Pande lab. It takes the middle path between a CPU's flexibility and a GPU's speed, performing a limited set of calculations rapidly while still retaining adaptable. However, unlike CPUs and GPUs, donors cannot perform other activities on their PS3 while running Folding@home. Instead, the Pande lab has specifically designed its Work Units to take approximately eight hours so that they can be completed overnight. The PS3's uniform console environment makes support
easier, as well as making Folding@home user friendly
. The PS3 also has the ability to stream data quickly to its GPU, allowing for real-time atomic detail visualizations of the protein dynamics.
(SMP) client fulfills two purposes: it takes advantage of the high-performance capabilities of recent multiprocessor
systems, and it helps develop a simulation architecture that will become one of the dominant FAH computing paradigms as multi-core chips become an industry standard over the next several years. The SMP client is capable of delivering over a 4x calculation speedup over the standard uniprocessor clients.
Folding@home's SMP core handles multi-core CPUs very different from other distributed computing projects, including those under BOINC. Instead of simply doing multiple Work Units simultaneously, single WUs are completed much faster across the multiple CPU cores. This cuts down on the traditional difficulties of scaling a large simulation to many processors. As such, this approach is very scientifically valuable. Some of the Pande lab publications would not have been possible without the SMP client.
On November 13, 2006, first generation SMP Folding@home clients for x86 Microsoft Windows
, x86-64
Linux
, and x86 Mac OS X
were released. These clients used Message Passing Interface
(MPI) protocols on the localhost
, as at the time the Gromacs cores were not designed to be used with multiple thread
s. This made Folding@home the first to use MPI for distributed computing software, as it had previously been reserved only for supercomputers. The MPI-based clients worked well in Unix
-based operating systems such as Linux
and Mac's OS-X
, but was particularly troublesome in Windows. Despite these difficulties, SMP1 generated significant results that would have been impossible otherwise and which represented a landmark in the simulation of protein folding.
The second generation of the SMP client was released as an open beta on January 24, 2010, and subsequently replaced SMP1. The SMP2 client exchanges the complex MPI for thread
s, which removes much of the overhead of keeping the cores synchronized. The SMP2 client also supports a bonus points system, which non-linearly rewards additional points to donors for quick WU returns and for contributing to next-generation capabilities. Donors who run the SMP2 client receive these extra points if they use a passkey and maintained an 80% successful return of Work Units.
SMP2 also supports extra-large Work Units for users with powerful eight-core CPUs or better. While these WUs consume even more RAM
and have more network usage than regular SMP WUs, users who run these are rewarded with a 20% increase over SMP2's bonus point system. These powerful computers allow for simulations to be performed on Folding@home that had previously required the use of supercomputing clusters. There is a great scientific need to run these simulations out to long timescales as quickly as possible, so the additional bonus points also serves as an incentive for rapid completions of Work Units. This allows the Pande lab to perform studies of larger molecular systems that would not have been possible anywhere else on Folding@home.
, Mac OS X
and Linux
operating systems. Following its predecessors, v7 runs Folding@home in the background at very low priority, which allows other applications to use CPU resources as they need. The v7 client is designed to make the installation, start-up, and operation user-friendly for novices, as well as offer greater scientific flexibility than previous clients. It is the Pande lab's goal to make v7 the recommended client by January 2012 at the latest, and versions of v7 will be frequently released until then.
V7 consists of several elements. The user interacts with v7's GUI
, known as FAHControl. It has Novice, Advanced, and Expert user interface modes, and has the ability to monitor, configure, and control many remote folding clients from a single computer. FAHControl can monitor and direct FAHClient, which runs behind the scenes and in turn manages each FAHSlot (or "slot"). These slots act as replacements for the previously distinct FAH clients, as they may be of Uniprocessor, SMP, or GPU type. Each slot also contains a core and data associated with it, and can download, process, and upload Work Units independently. The FAHViewer function, modeled after the PS3 viewer, displays a real-time 3D rendering, if available, of the protein currently being processed.
s to retrieve a Work Unit, processes it, and returns it upon completion. During transfer, all Work Units are validated through the use of 2048-bit digital signature
s. These WUs have associated deadlines and credit (point) value. If this deadline is exceeded, the user may not get credit and the unit will be reissued to someone else. As protein folding is serial in nature and each WU is generated from its predecessor, this allows the overall simulation process to proceed normally if a WU is not returned after a certain period of time. Due to these deadlines, the minimum system requirements for Folding@home is a Pentium 3 450 MHz CPU with SSE
or newer. However, Work Units for high performance clients have a much shorter deadline than those for the uniprocessor client, as a major part of the scientific benefit is dependent on rapidly completing simulations.
Before public release, Work Units go through several Quality Assurance
steps to keep problematic WUs from becoming fully available. But unlike particular BOINC projects such as SETI@home
, Folding@home's Work Units are normally processed only once, except in the rare event that errors occur during processing of a WU. If this occurs for three different donors, it is automatically pulled from distribution. Topics in the Folding@home forum can be used to differentiate between problematic hardware and an actual bad Work Unit.
Work Units are very much tied to the Pande lab's simulation Markov state models
, which allow for extensive parallelization
of very long simulation processes which otherwise seem intrinsically serial. During the folding process, proteins spend much of their time "waiting" in various states, before quickly transitioning to the next configuration. This allows for the unique possibility to simulate only a small fraction of the overall folding timescale, leading to significant speedups. The Pande lab achieves this by first dividing the protein's possible dynamics into a series of related conformation states, and creates WUs to calculate the rates of transition between these states. When the completed WUs are gathered, the Pande lab then runs sophisticated Bayesian Machine Learning
algorithms which calculate which states are reasonable and the rates between them. This also averages
the molecular simulation ensembles, which is important so that direct, meaningful comparison between in silico simulations and in vitro experiments can be made. This system is successful even at the millisecond timescale, compares well to traditional methods and experimental results, and allows for previously intractable problems to be within reach.
programs, including GROMACS, AMBER
, TINKER
, CPMD
, SHARPEN, ProtoMol, BrookGPU
and Desmond
. Some of these cores perform explicit atom-by-atom molecular dynamics
calculations, while others perform implicit solvation
methods, which treat atoms as a mathematical continuum. These cores are open-source software
or are under similar licenses, and are verified during download by 2048-bit digital signatures. While the same core can be used by various versions of the client, separating the core from the client enables the scientific methods to be updated automatically as needed without a client update.
is a distributed computing project aimed at protein structure prediction
and is one of the most successful approaches to this problem. Folding@home and Rosetta@home address very different molecular questions. Although Rosetta@home does not provide information into how proteins fold, it does predict the protein's most likely final structure, which in some cases is used as a basis for Folding@home's projects. Rosetta's predictions can help FAH simulate the folding of larger proteins more efficiently. Folding@home can also verify Rosetta@home's results and find additional atomistic details of the protein's kinetics and folding pathway, which is intrinsically much more difficult. Folding@home's accurate simulations have also suggesting important novel implications into the fields of protein folding, structure prediction, and certain folding experiments, and have shown that Rosetta's structure prediction may benefit from thermodynamic sampling aspects of protein folding mechanisms.
Folding@home also compares well to Anton
, a powerful supercomputer which uses specialized hardware to produce a small number of ultra-long molecular trajectories. It is unique in this ability, and like Folding@home, has also improved particular long-held theories of protein folding. Its longer simulations, while computationally expensive, contain more phase space
than any one of Folding@home's many shorter trajectories, which allows Anton to perform a thorough exploration of the required space. As of October 2011, Anton and FAH are the two most powerful molecular dynamics systems, and Anton has also run simulations out to the millisecond range. In 2011, the Pande lab built a Markov state model from one of Anton's simulations. It demonstrated that there was little difference between MSMs built from Anton's fewer long trajectories and one assembled from Folding@home's many shorter trajectories. Their analysis also showed that Folding@home's Markov state models significantly improve the analysis of these longer simulations, such as revealing additional relevant folding pathways and information into how the protein carries out its biological function. Folding@home is running further analysis on one of Anton's simulations to better determine how its approaches compare to Anton's methods. It is probable that a combination of Anton's and FAH's simulation methods would be very beneficial, and Pande looks forward to see how Anton and FAH can be used together.
Distributed computing
Distributed computing is a field of computer science that studies distributed systems. A distributed system consists of multiple autonomous computers that communicate through a computer network. The computers interact with each other in order to achieve a common goal...
project designed to use spare processing power on personal computers to perform simulations of disease-relevant protein folding
Protein folding
Protein folding is the process by which a protein structure assumes its functional shape or conformation. It is the physical process by which a polypeptide folds into its characteristic and functional three-dimensional structure from random coil....
and other molecular dynamics
Molecular dynamics
Molecular dynamics is a computer simulation of physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a period of time, giving a view of the motion of the atoms...
, and to improve on the methods of doing so. Also referred to as FAH or F@h, much of its work attempts to determine how proteins reach their final structure, which is of significant academic interest and has implications to both disease research and nanotechnology
Nanotechnology
Nanotechnology is the study of manipulating matter on an atomic and molecular scale. Generally, nanotechnology deals with developing materials, devices, or other structures possessing at least one dimension sized from 1 to 100 nanometres...
. To a lesser degree Folding@home also tries to predict
Protein structure prediction
Protein structure prediction is the prediction of the three-dimensional structure of a protein from its amino acid sequence — that is, the prediction of its secondary, tertiary, and quaternary structure from its primary structure. Structure prediction is fundamentally different from the inverse...
that final structure from only the initial amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
sequence, which has applications in drug design
Drug design
Drug design, also sometimes referred to as rational drug design or structure-based drug design, is the inventive process of finding new medications based on the knowledge of the biological target...
. Folding@home is developed and operated by the Pande Laboratory at Stanford University
Stanford University
The Leland Stanford Junior University, commonly referred to as Stanford University or Stanford, is a private research university on an campus located near Palo Alto, California. It is situated in the northwestern Santa Clara Valley on the San Francisco Peninsula, approximately northwest of San...
, under the direction of Vijay Pande. The goal of the project is to "understand protein folding, misfolding, and related diseases".
Folding@home's simulations of protein folding and misfolding enable the scientific community to better understand the development of many diseases, including Alzheimer's disease
Alzheimer's disease
Alzheimer's disease also known in medical literature as Alzheimer disease is the most common form of dementia. There is no cure for the disease, which worsens as it progresses, and eventually leads to death...
, Parkinson's disease
Parkinson's disease
Parkinson's disease is a degenerative disorder of the central nervous system...
, cancer
Cancer
Cancer , known medically as a malignant neoplasm, is a large group of different diseases, all involving unregulated cell growth. In cancer, cells divide and grow uncontrollably, forming malignant tumors, and invade nearby parts of the body. The cancer may also spread to more distant parts of the...
, Creutzfeldt–Jakob disease, Huntington's disease
Huntington's disease
Huntington's disease, chorea, or disorder , is a neurodegenerative genetic disorder that affects muscle coordination and leads to cognitive decline and dementia. It typically becomes noticeable in middle age. HD is the most common genetic cause of abnormal involuntary writhing movements called chorea...
, cystic fibrosis
Cystic fibrosis
Cystic fibrosis is a recessive genetic disease affecting most critically the lungs, and also the pancreas, liver, and intestine...
, sickle-cell anaemia, HIV
HIV
Human immunodeficiency virus is a lentivirus that causes acquired immunodeficiency syndrome , a condition in humans in which progressive failure of the immune system allows life-threatening opportunistic infections and cancers to thrive...
, Chagas disease
Chagas disease
Chagas disease is a tropical parasitic disease caused by the flagellate protozoan Trypanosoma cruzi. T. cruzi is commonly transmitted to humans and other mammals by an insect vector, the blood-sucking insects of the subfamily Triatominae most commonly species belonging to the Triatoma, Rhodnius,...
, influenza
Influenza
Influenza, commonly referred to as the flu, is an infectious disease caused by RNA viruses of the family Orthomyxoviridae , that affects birds and mammals...
, osteogenesis imperfecta
Osteogenesis imperfecta
Osteogenesis imperfecta is a genetic bone disorder. People with OI are born with defective connective tissue, or without the ability to make it, usually because of a deficiency of Type-I collagen...
, autism
Autism
Autism is a disorder of neural development characterized by impaired social interaction and communication, and by restricted and repetitive behavior. These signs all begin before a child is three years old. Autism affects information processing in the brain by altering how nerve cells and their...
, and alpha 1-antitrypsin deficiency
Alpha 1-antitrypsin deficiency
Alpha 1-antitrypsin deficiency is an autosomal recessive genetic disorder caused by defective production of alpha 1-antitrypsin , leading to decreased A1AT activity in the blood and lungs, and deposition of excessive abnormal A1AT protein in liver cells...
, among others. More fundamentally, understanding the process of protein folding — how proteins assemble themselves into a functional state — is one of the outstanding problems of molecular biology
Molecular biology
Molecular biology is the branch of biology that deals with the molecular basis of biological activity. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry...
. The Pande lab has produced ninety-five scientific research papers
Academic publishing
Academic publishing describes the subfield of publishing which distributes academic research and scholarship. Most academic work is published in journal article, book or thesis form. The part of academic written output that is not formally published but merely printed up or posted is often called...
as a direct result of the project, and Folding@home has caused paradigm shift
Paradigm shift
A Paradigm shift is, according to Thomas Kuhn in his influential book The Structure of Scientific Revolutions , a change in the basic assumptions, or paradigms, within the ruling theory of science...
s in protein folding theory.
In January 2010 the Folding@home project successfully simulated protein folding in the 1.5 millisecond
Millisecond
A millisecond is a thousandth of a second.10 milliseconds are called a centisecond....
range — a simulation thousands of times longer than ever previously achieved. Folding@home has pioneered the uses of GPUs, multi-core processors, and PlayStation 3
PlayStation 3
The is the third home video game console produced by Sony Computer Entertainment and the successor to the PlayStation 2 as part of the PlayStation series. The PlayStation 3 competes with Microsoft's Xbox 360 and Nintendo's Wii as part of the seventh generation of video game consoles...
s for distributed computing. It remains one of the world's fastest computing systems, and is more powerful than all distributed computing projects under BOINC combined. Folding@home's distributed simulations remain accurate compared to results from laboratory research, a "grand challenge" in computational biology
Computational biology
Computational biology involves the development and application of data-analytical and theoretical methods, mathematical modeling and computational simulation techniques to the study of biological, behavioral, and social systems...
. The Pande lab's goal is to refine Folding@home's methods to the level where it will become an essential tool for molecular medical research, and they collaborate with various scientific institutions and laboratories across the world.
Biomedical significance

Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s are an essential component to many biological functions, and participate in virtually every process within cells. They often act as enzymes, performing biochemical reactions including cell signaling
Cell signaling
Cell signaling is part of a complex system of communication that governs basic cellular activities and coordinates cell actions. The ability of cells to perceive and correctly respond to their microenvironment is the basis of development, tissue repair, and immunity as well as normal tissue...
, transportation, cellular regulation, and others. As structural elements, some proteins act as a type of skeleton for cells
Cytoskeleton
The cytoskeleton is a cellular "scaffolding" or "skeleton" contained within a cell's cytoplasm and is made out of protein. The cytoskeleton is present in all cells; it was once thought to be unique to eukaryotes, but recent research has identified the prokaryotic cytoskeleton...
, and as antibodies other proteins help the immune system
Immune system
An immune system is a system of biological structures and processes within an organism that protects against disease by identifying and killing pathogens and tumor cells. It detects a wide variety of agents, from viruses to parasitic worms, and needs to distinguish them from the organism's own...
. Before a protein can take on these roles, it must fold into a functional three-dimensional shape based on a particular series of steps
Reaction intermediate
A reaction intermediate or an intermediate is a molecular entity that is formed from the reactants and reacts further to give the directly observed products of a chemical reaction. Most chemical reactions are stepwise, that is they take more than one elementary step to complete...
, which often occur spontaneously
Spontaneous process
A spontaneous process is the time-evolution of a system in which it releases free energy and moves to a lower, more thermodynamically stable energy state...
. Understanding protein folding is thus critical to understanding what a protein does and how it works. Moreover, when proteins do not fold correctly—that is, when they fold down the wrong pathway and end up misshapen—they can aggregate
Protein aggregation
Protein aggregation is the aggregation of mis-folded proteins, and is thought to be responsible for many degenerative diseases, such as Alzheimer's. It has also been implicated in CAG repeat diseases....
and cause serious and in some cases life-threatening diseases. However, once the misfolding process is determined, figuring out its prevention can be the next step, and preventative treatments can be applied during the folding process. While the understanding of protein folding requires a combination of theories and experiments, creating experimentally comparable simulations of protein folding dynamics remains difficult and is considered a "holy grail" of computational biology
Computational biology
Computational biology involves the development and application of data-analytical and theoretical methods, mathematical modeling and computational simulation techniques to the study of biological, behavioral, and social systems...
. Folding@home's distributed computing methods can address these challenges and allow for unique insights into the complexity of protein folding while strongly agreeing with experimental data.
Since the late 1990s, simulations of protein molecular dynamics have been severely limited by computational power. Straightforward approaches have exceptional difficulty for all but the most elementary of systems, which has prompted the use of simplified native
Native state
In biochemistry, the native state of a protein is its operative or functional form. While all protein molecules begin as simple unbranched chains of amino acids, once completed they assume highly specific three-dimensional shapes; that ultimate shape, known as tertiary structure, is the folded...
-centric in silico
In silico
In silico is an expression used to mean "performed on computer or via computer simulation." The phrase was coined in 1989 as an analogy to the Latin phrases in vivo and in vitro which are commonly used in biology and refer to experiments done in living organisms and outside of living organisms,...
molecular models. Not only are these models insufficient for a comprehensive view of protein folding, but the traditional methods of using a small number of very long simulations cannot accurately capture enough detailed information into how a protein misfolds. Moreover, these simplified models often ignore how solvent
Solvent
A solvent is a liquid, solid, or gas that dissolves another solid, liquid, or gaseous solute, resulting in a solution that is soluble in a certain volume of solvent at a specified temperature...
s such as water might affect the folding process. In strong agreement with experimental observations, its influence was revealed in 2004 using Folding@home's computational power and explicit solvation simulations. This observation was made possible by Folding@home's statistical assembly of many shorter simulations, which compared to traditional approaches, provide a much more complete description of the protein's energy landscape
Energy landscape
In physics, an energy landscape is a mapping of all possible conformations of a molecular entity, or the spatial positions of interacting molecules in a system, and their corresponding energy levels, typically Gibbs free energy, on a two- or three-dimensional Cartesian coordinate system.In...
, conformation space
Configuration space
- Configuration space in physics :In classical mechanics, the configuration space is the space of possible positions that a physical system may attain, possibly subject to external constraints...
, and equilibrium thermodynamics
Equilibrium thermodynamics
Equilibrium Thermodynamics is the systematic study of transformations of matter and energy in systems as they approach equilibrium. The word equilibrium implies a state of balance. Equilibrium thermodynamics, in origins, derives from analysis of the Carnot cycle. Here, typically a system, as...
. While some of these simulations may work themselves into impossible atomic configurations or into the correct native state, others will illustrate how that protein misfolds. By utilizing thermodynamics
Thermodynamics
Thermodynamics is a physical science that studies the effects on material bodies, and on radiation in regions of space, of transfer of heat and of work done on or by the bodies or radiation...
, Folding@home uses methods that can find these rare events without having to complete many simulations. This allows for a highly accurate and thorough exploration of protein configurations in a reasonable amount of time, even over very long simulation timescales. For the instrumental development of the software behind these statistical approaches and for attaining quantitative agreement between theory and experiment, in 2010 Folding@home researcher Greg Bowman was awarded the Thomas Kuhn
Thomas Kuhn
Thomas Samuel Kuhn was an American historian and philosopher of science whose controversial 1962 book The Structure of Scientific Revolutions was deeply influential in both academic and popular circles, introducing the term "paradigm shift," which has since become an English-language staple.Kuhn...
Paradigm Shift Award from the American Chemical Society
American Chemical Society
The American Chemical Society is a scientific society based in the United States that supports scientific inquiry in the field of chemistry. Founded in 1876 at New York University, the ACS currently has more than 161,000 members at all degree-levels and in all fields of chemistry, chemical...
.
The Pande lab and other researchers can use Folding@home to study aspects of folding, misfolding, and related diseases that would never be seen experimentally. For example, most proteins have such intrinsically stable native state
Native state
In biochemistry, the native state of a protein is its operative or functional form. While all protein molecules begin as simple unbranched chains of amino acids, once completed they assume highly specific three-dimensional shapes; that ultimate shape, known as tertiary structure, is the folded...
s that it is difficult to experimentally study their folding events, and researchers must resort to chemical denaturation
Denaturation (biochemistry)
Denaturation is a process in which proteins or nucleic acids lose their tertiary structure and secondary structure by application of some external stress or compound, such as a strong acid or base, a concentrated inorganic salt, an organic solvent , or heat...
methods. While the denatured state of proteins is of scientific interest because any residual structure may affect its folding behavior, even with molecular experiments it remains difficult to determine the extent of a local structure that may be present. However, molecular simulations on Folding@home can provide detail into denatured conformational states and insights into the chemical denaturation mechanisms. These and other simulations run on Folding@home are used in conjunction with laboratory experiments. Additionally, as of 2011 the Pande lab is performing studies into how protein folding in their native cells
Cell (biology)
The cell is the basic structural and functional unit of all known living organisms. It is the smallest unit of life that is classified as a living thing, and is often called the building block of life. The Alberts text discusses how the "cellular building blocks" move to shape developing embryos....
may be different than in environments such as test tube
Test tube
A test tube, also known as a culture tube or sample tube, is a common piece of laboratory glassware consisting of a finger-like length of glass or clear plastic tubing, open at the top, usually with a rounded U-shaped bottom....
s used during experiments.
In addition to the diseases listed below, researchers use Folding@home to study malaria
Malaria
Malaria is a mosquito-borne infectious disease of humans and other animals caused by eukaryotic protists of the genus Plasmodium. The disease results from the multiplication of Plasmodium parasites within red blood cells, causing symptoms that typically include fever and headache, in severe cases...
, Chagas disease
Chagas disease
Chagas disease is a tropical parasitic disease caused by the flagellate protozoan Trypanosoma cruzi. T. cruzi is commonly transmitted to humans and other mammals by an insect vector, the blood-sucking insects of the subfamily Triatominae most commonly species belonging to the Triatoma, Rhodnius,...
, the prion
Prion
A prion is an infectious agent composed of protein in a misfolded form. This is in contrast to all other known infectious agents which must contain nucleic acids . The word prion, coined in 1982 by Stanley B. Prusiner, is a portmanteau derived from the words protein and infection...
s which cause Creutzfeldt–Jakob disease, and details into how virus
Virus
A virus is a small infectious agent that can replicate only inside the living cells of organisms. Viruses infect all types of organisms, from animals and plants to bacteria and archaea...
es such as HIV
HIV
Human immunodeficiency virus is a lentivirus that causes acquired immunodeficiency syndrome , a condition in humans in which progressive failure of the immune system allows life-threatening opportunistic infections and cancers to thrive...
and influenza
Influenza
Influenza, commonly referred to as the flu, is an infectious disease caused by RNA viruses of the family Orthomyxoviridae , that affects birds and mammals...
function and infect cellular membranes. Results from this research have led to major shifts in the understanding of protein folding and its applications for disease, as well as improved protein folding models. Folding@home is dedicated to producing significant amounts of results towards protein folding, the diseases that result from protein misfolding, and novel computational methods for doing so. The goal of the first five years of the project was to make significant advances in understanding folding, while the current goal is to understand misfolding and related disease, especially Alzheimer's disease.
As a part of Stanford University, a non-profit organization, the Pande lab does not sell the results generated by Folding@home. The large data sets from the project are freely available for other researchers to use upon request, and some can be accessed from the Folding@home website. The Pande lab also releases Folding@home's key software to other researchers, so that the algorithms which benefit Folding@home will also aid other scientific areas. Moreover, in 2011 they released the open-source Copernicus software, so that other researchers can run molecular simulations much more efficiently on clusters or supercomputers. Summaries of all of the scientific findings from Folding@home are posted on the Folding@home website after publication. The full publications are available online or from a local municipal or academic library.
Alzheimer's disease
Alzheimer's diseaseAlzheimer's disease
Alzheimer's disease also known in medical literature as Alzheimer disease is the most common form of dementia. There is no cure for the disease, which worsens as it progresses, and eventually leads to death...
, a form of dementia
Dementia
Dementia is a serious loss of cognitive ability in a previously unimpaired person, beyond what might be expected from normal aging...
which most often affects the elderly, is believed to be caused by specific misfolding and subsequent aggregation
Protein aggregation
Protein aggregation is the aggregation of mis-folded proteins, and is thought to be responsible for many degenerative diseases, such as Alzheimer's. It has also been implicated in CAG repeat diseases....
of the small 42-residue
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
amyloid beta
Amyloid beta
Amyloid beta is a peptide of 36–43 amino acids that is processed from the Amyloid precursor protein. While it is most commonly known in association with Alzheimer's disease, it does not exist specifically to cause disease...
(Aß) peptide
Peptide
Peptides are short polymers of amino acid monomers linked by peptide bonds. They are distinguished from proteins on the basis of size, typically containing less than 50 monomer units. The shortest peptides are dipeptides, consisting of two amino acids joined by a single peptide bond...
. The severity of the disease depends not only on the amount of Aß, but also on how it misfolds. Current theory holds that toxic non-plaque
Senile plaques
Senile plaques are extracellular deposits of amyloid in the gray matter of the brain. The deposits are associated with degenerative neural structures and an abundance of microglia and astrocytes...
Aß oligomers (aggregates of many monomers) bind to a surface receptor on neuron
Neuron
A neuron is an electrically excitable cell that processes and transmits information by electrical and chemical signaling. Chemical signaling occurs via synapses, specialized connections with other cells. Neurons connect to each other to form networks. Neurons are the core components of the nervous...
s and change the structure of the synapse
Synapse
In the nervous system, a synapse is a structure that permits a neuron to pass an electrical or chemical signal to another cell...
, thereby disrupting neuronal communication and causing neuronal cell death which leads to the associated neurodegenerative consequences. Understanding how and why this peptide misfolds could result in key insights into how to cure Alzheimer's Disease, and will also help the Pande lab prepare for similar aggregation studies.
Despite this connection, toxic Aß aggregations remain so complex that it was not previously possible to simulate them in atomic resolution. In 2011, the Pande lab explored how their Abeta studies using Folding@home could be used as a starting point for a new Alzheimer's therapy. Folding@home is currently concentrating on Alzheimer's and continues full-scale simulations of amyloid beta and its oligomerization, which had previously been a technological challenge to simulate. These studies build off of the Pande lab's 2008 published work into new ways to simulate Abeta oligomerization over long timescales. In the same publication, previous all-atom simulations were performed that led to specific experimentally-tested predictions, such as ways in which to stabilize the protein and prevent the toxic oligomer formation. The Pande lab is focusing their research in this area for rational drug design
Drug design
Drug design, also sometimes referred to as rational drug design or structure-based drug design, is the inventive process of finding new medications based on the knowledge of the biological target...
approaches. Pande described that paper as the "tip of the iceberg" for the Folding@home studies of Alzheimer's, as further results will follow and possibly new therapeutics.
Folding@home is also being used to study Aß fragments of different sizes to determine how various natural enzymes affect the structure and folding of Aß. These fragments are tied to senile plaques, a pathological marker of Alzheimer's disease in patient's brain. When certain enzymes cleave the amyloid precursor protein
Amyloid precursor protein
Amyloid precursor protein is an integral membrane protein expressed in many tissues and concentrated in the synapses of neurons. Its primary function is not known, though it has been implicated as a regulator of synapse formation, neural plasticity and iron export...
, Abeta peptides are produced, while the action of other enzymes can instead produce p3 peptides, much smaller fragments of Aß. Folding@home is simulating one of these smaller peptides in water in an effort to determine how the length of Aß affects its overall structure.
In 2010, several possible drug leads predicted by Folding@home went from the test tube
Test tube
A test tube, also known as a culture tube or sample tube, is a common piece of laboratory glassware consisting of a finger-like length of glass or clear plastic tubing, open at the top, usually with a rounded U-shaped bottom....
to testing on living tissue
Tissue (biology)
Tissue is a cellular organizational level intermediate between cells and a complete organism. A tissue is an ensemble of cells, not necessarily identical, but from the same origin, that together carry out a specific function. These are called tissues because of their identical functioning...
, and in close cooperation with the Nanomedicine Center for Protein Folding, the drug leads continued to be refined. Additionally, as predicted by FAH's simulations, a stable form of amyloid beta was experimentally verified which the Pande lab believes could be used as a starting point for new Alzheimer's therapy. In 2008, Folding@home produced several small drug candidates to fight Alzheimer's Disease, as they appear to inhibit the toxicity of Abeta.
The Pande lab is also using Folding@home to investigate protein–protein interactions, which occur extensively throughout both benign and disease-related biological activities. Interactions involving the common SH3
SH3 domain
The SRC Homology 3 Domain is a small protein domain of about 60 amino acids residues first identified as a conserved sequence in the viral adaptor protein v-Crk and the non-catalytic parts of enzymes such as phospholipase and several cytoplasmic tyrosine kinases such as Abl and Src...
protein are also being studied, as it has implications in Alzheimers research. The refinement of these simulations has greatly improved the Pande lab's ability to understand a wide variety of biological interactions.
Huntington's disease
Huntington's diseaseHuntington's disease
Huntington's disease, chorea, or disorder , is a neurodegenerative genetic disorder that affects muscle coordination and leads to cognitive decline and dementia. It typically becomes noticeable in middle age. HD is the most common genetic cause of abnormal involuntary writhing movements called chorea...
, an incurable neurodegenerative genetic disorder affecting muscle coordination
Motor coordination
thumb|right|Motor coordination is shown in this animated sequence by [[Eadweard Muybridge]] of himself throwing a diskMotor coordination is the combination of body movements created with the kinematic and kinetic parameters that result in intended actions. Such movements usually smoothly and...
and leading to dementia
Dementia
Dementia is a serious loss of cognitive ability in a previously unimpaired person, beyond what might be expected from normal aging...
, is also associated with protein misfolding. Specifically, it is caused by a mutation in the Huntingtin gene, which causes excessively long repetitive chains
Polyglutamine tract
A polyglutamine tract or polyQ tract is a portion of a protein consisting of a sequence of several glutamine units. A tract typically consists of about 10 to a few hundred such units....
of the glutamine
Glutamine
Glutamine is one of the 20 amino acids encoded by the standard genetic code. It is not recognized as an essential amino acid but may become conditionally essential in certain situations, including intensive athletic training or certain gastrointestinal disorders...
amino acid in the Huntingtin protein, a protein that plays important roles in nerve cells. The likelihood of neuronal cell death is primarily affected by the length of the glutamine chain and the neuron's intracellular exposure to the misfolded Huntingtin protein. The defective protein causes Huntington's by aggregating most often in the striatum
Striatum
The striatum, also known as the neostriatum or striate nucleus, is a subcortical part of the forebrain. It is the major input station of the basal ganglia system. The striatum, in turn, gets input from the cerebral cortex...
and frontal cortex of patient's brains. The Pande lab is using Folding@home to study these aggregates, as well as predict how they form. How this aggregation occurs has been largely unknown, but in 2009 a paper based off of Folding@home's results and published in the Journal of Molecular Biology
Journal of Molecular Biology
The Journal of Molecular Biology is a peer-reviewed scientific journal published weekly by Elsevier. It covers original scientific research concerning studies of organisms or their components at the molecular level.- Notable articles :...
investigated possible mechanisms for the aggregation formation, and the implications into how to prevent it. These studies will be useful for drug design
Drug design
Drug design, also sometimes referred to as rational drug design or structure-based drug design, is the inventive process of finding new medications based on the knowledge of the biological target...
approaches against the disease, and will serve as a foundation for methods to stop the aggregation formation. Additionally, some of the methods used to study Huntington's are also being used for Alzheimer's research.
In 2010, Folding@home researcher Veena Thomas proposed a novel therapeutic strategy for HD, which may be funded by the NIH
National Institutes of Health
The National Institutes of Health are an agency of the United States Department of Health and Human Services and are the primary agency of the United States government responsible for biomedical and health-related research. Its science and engineering counterpart is the National Science Foundation...
. This strategy could be used to bring the results from Folding@home directly to a therapeutic.
Cancer
More than half of all known cancerCancer
Cancer , known medically as a malignant neoplasm, is a large group of different diseases, all involving unregulated cell growth. In cancer, cells divide and grow uncontrollably, forming malignant tumors, and invade nearby parts of the body. The cancer may also spread to more distant parts of the...
s involve mutations in p53
P53
p53 , is a tumor suppressor protein that in humans is encoded by the TP53 gene. p53 is crucial in multicellular organisms, where it regulates the cell cycle and, thus, functions as a tumor suppressor that is involved in preventing cancer...
, a tumor suppressor protein present in every cell which signals for cell death in the event of damage to a cell's DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
. If p53 becomes mutated, breaks down, or fails to fold properly, it will no longer be able to cause the "stop signal" for cell division. Cells will consequently divide and grow uncontrollably to form tumors. Folding@home is used to study specific properties of p53 in order to understand and predict these mutations, and in 2007 work began to develop inhibitor proteins that deactivate damaged p53. The Pande lab has also developed a novel experimentally-verified method for predicting how mutations affect p53, which has had reasonable success in the identification of deleterious mutations such as those linked to cancer. This supplemented work performed in 2005 that studied how mutations affect the folding of p53, and which mutations are relevant to cancer. While it was the first peer-reviewed publication from a distributed computing project related to cancer, the study also agreed well with experiments and offered insights that were previously unobtainable. Following these results, the Pande lab have expanded their work to other p53-related diseases.
The Pande lab is also performing research into protein chaperones. These are proteins that assist in the folding of other molecules, assembly of oligomeric structures, the prevention of potential damage caused by protein misfolding, and other functions. They are needed for these purposes by rapidly growing cancerous cells. Using Folding@home and working closely with the Protein Folding Center, they plan to find ways to inhibit chaperones involved in cancer. Using Folding@home for a more comprehensive visualization of their functions, the Pande lab and the Protein Folding Center collectively plan to engineer modified chaperonins to inhibit the folding of particular proteins associated with human diseases such as cancer and Alzheimer's. While this approach has been used before, they believe that this project, if successful, could lead to an interesting new drug against cancer or at least make major advances in that area.
Folding@home is also used to study the folding of several other proteins which have mutations tied to cancer, such as the enzyme src Kinase and certain forms of the Engrailed
Engrailed (gene)
engrailed is a homeodomain transcription factor involved in many aspects of multicellular development. First known for its role in arthropod embryological development, working in consort with the Hox genes, engrailed has been found to be important in other areas of development...
Homeodomain. These proteins also have a great deal of experimental data for comparison, and serve as a great system for the understanding of folding and misfolding. Additionally, the Pande lab is using Folding@home to understand the dynamics of a small knottin
Trefoil knot fold
The trefoil knot fold is a protein fold in which the protein backbone is twisted into a trefoil knot shape. "Shallow" knots in which the tail of the polypeptide chain only passes through a loop by a few residues are uncommon, but "deep" knots in which many residues are passed through the loop are...
protein and how it can be used to bind to contrast agents for imaging scan
Medical imaging
Medical imaging is the technique and process used to create images of the human body for clinical purposes or medical science...
or drug
Drug
A drug, broadly speaking, is any substance that, when absorbed into the body of a living organism, alters normal bodily function. There is no single, precise definition, as there are different meanings in drug control law, government regulations, medicine, and colloquial usage.In pharmacology, a...
s. Finally, some forms of interleukin-2, an important signaling protein for the immune system, have been used as immunotherapy
Immunotherapy
Immunotherapy is a medical term defined as the "treatment of disease by inducing, enhancing, or suppressing an immune response". Immunotherapies designed to elicit or amplify an immune response are classified as activation immunotherapies. While immunotherapies that reduce or suppress are...
for cancer. The Pande lab believes that Folding@home's simulations of its dynamics will lead to insights into how to design other therapeutics.
Parkinson's disease
Parkinson's diseaseParkinson's disease
Parkinson's disease is a degenerative disorder of the central nervous system...
is a degenerative
Neurodegeneration
Neurodegeneration is the umbrella term for the progressive loss of structure or function of neurons, including death of neurons. Many neurodegenerative diseases including Parkinson’s, Alzheimer’s, and Huntington’s occur as a result of neurodegenerative processes. As research progresses, many...
disorder of the central nervous system
Central nervous system
The central nervous system is the part of the nervous system that integrates the information that it receives from, and coordinates the activity of, all parts of the bodies of bilaterian animals—that is, all multicellular animals except sponges and radially symmetric animals such as jellyfish...
, characterized by shaking, rigidity, slowness of movement, and dementia
Dementia
Dementia is a serious loss of cognitive ability in a previously unimpaired person, beyond what might be expected from normal aging...
. The Pande lab has performed preliminary studies on the properties of alpha-synuclein
Alpha-synuclein
Alpha-synuclein is a protein that, in humans, is encoded by the SNCA gene. An alpha-synuclein fragment, known as the non-Abeta component of Alzheimer's disease amyloid, originally found in an amyloid-enriched fraction, is shown to be a fragment of its precursor protein, NACP, by cloning of the...
, a key natively unfolded protein
Intrinsically unstructured proteins
Intrinsically unstructured proteins, often referred to as naturally unfolded proteins or disordered proteins, are proteins characterized by lack of stable tertiary structure when the protein exists as an isolated polypeptide chain under physiological conditions in vitro...
. Particular mutations of alpha-synuclein can aggregate to form toxic fibrils, and while the mechanism of this aggregation remains largely unknown, it can lead to the Parkinson's disease and other conditions. The Pande lab is also testing how Folding@home's methods apply to this problem, and in 2005, Pande presented results from FAH at a National Parkinson Foundation
National Parkinson Foundation
The National Parkinson Foundation , founded in 1957, is a national organization whose mission is to improve the quality of care for people with Parkinson's through clinical research, education and outreach on Parkinson’s disease....
conference.
Osteogenesis imperfecta
Osteogenesis imperfectaOsteogenesis imperfecta
Osteogenesis imperfecta is a genetic bone disorder. People with OI are born with defective connective tissue, or without the ability to make it, usually because of a deficiency of Type-I collagen...
is a non-curable genetic bone disorder. Those with the disease are unable to successfully make functional connective bone tissue. This is lethal for many but can also induce a higher rate of miscarriages. The disease is caused by mutations in the Type-1 collagen
Collagen
Collagen is a group of naturally occurring proteins found in animals, especially in the flesh and connective tissues of mammals. It is the main component of connective tissue, and is the most abundant protein in mammals, making up about 25% to 35% of the whole-body protein content...
protein, the most common form of collagen and found abundantly throughout the body. Although some of these mutations of collagen can lead to serious morphological disorders, more benign forms can cause brittle bones and other subtleties. Folding@home has performed simulations of collagen, and has produced a paper on Osteogenesis imperfecta outlining new molecular simulation techniques and revealing new insights into how collagen misfolds. The Pande lab believes these results will be useful for later computational studies of collagen.
Diabetes
AmylinAmylin
Amylin, or Islet Amyloid Polypeptide , is a 37-residue peptide hormone secreted by pancreatic β-cells at the same time as insulin .-Clinical significance:...
is a misfolded peptide
Peptide
Peptides are short polymers of amino acid monomers linked by peptide bonds. They are distinguished from proteins on the basis of size, typically containing less than 50 monomer units. The shortest peptides are dipeptides, consisting of two amino acids joined by a single peptide bond...
involved in Type II diabetes. While amylin is natively unfolded, it forms an alpha helix
Alpha helix
A common motif in the secondary structure of proteins, the alpha helix is a right-handed coiled or spiral conformation, in which every backbone N-H group donates a hydrogen bond to the backbone C=O group of the amino acid four residues earlier...
structure upon contact with cellular membranes. Moreover, it can aggregate into large deposits on these membranes, inducing cell death of insulin
Insulin
Insulin is a hormone central to regulating carbohydrate and fat metabolism in the body. Insulin causes cells in the liver, muscle, and fat tissue to take up glucose from the blood, storing it as glycogen in the liver and muscle....
-producing cells, which may be relevant to the development of the disease. Around 95% of patients with Type II diabetes exhibit these aggregate deposits. As of 2011, Folding@home is simulating amylin with the goal to understand how this aggregation forms and to design drugs to prevent it.
Antibiotics
The ribosomeRibosome
A ribosome is a component of cells that assembles the twenty specific amino acid molecules to form the particular protein molecule determined by the nucleotide sequence of an RNA molecule....
is a large biological machine that synthesizes proteins from mRNA. It is the target for approximately half of all known antibiotics, which usually kill bacteria by preventing their ribosomes from making new and essential proteins. Folding@home is simulating the ribosome in detail using new state-of-the-art calculation methods. Results from these simulations have significantly helped the Pande lab prepare to study more complex biomedical problems. The Pande lab is also using Folding@home to perform antibiotic drug design calculations.
Drug design
The Pande lab is using Folding@home to explore how to model and accurately estimate the binding energyBinding energy
Binding energy is the mechanical energy required to disassemble a whole into separate parts. A bound system typically has a lower potential energy than its constituent parts; this is what keeps the system together—often this means that energy is released upon the creation of a bound state...
of small molecules to a protein. Accurate predictions of binding affinities have the potential to significantly lower the development cost of new drugs. Additionally, Folding@home is utilized to find prime binding sites on protein surfaces by simulating interactions between ligand binding sites with different molecules. This has a direct application to computational drug design
Drug design
Drug design, also sometimes referred to as rational drug design or structure-based drug design, is the inventive process of finding new medications based on the knowledge of the biological target...
. Folding@home is also performing calculations on beta-lactamase
Beta-lactamase
Beta-lactamases are enzymes produced by some bacteria and are responsible for their resistance to beta-lactam antibiotics like penicillins, cephamycins, and carbapenems . These antibiotics have a common element in their molecular structure: a four-atom ring known as a beta-lactam...
, a protein that plays important roles in drug resistance
Drug resistance
Drug resistance is the reduction in effectiveness of a drug such as an antimicrobial or an antineoplastic in curing a disease or condition. When the drug is not intended to kill or inhibit a pathogen, then the term is equivalent to dosage failure or drug tolerance. More commonly, the term is used...
. The Pande lab hopes that by understanding its dynamics, they may be able to design drugs to deactivate it.
Participation

PlayStation 3
The is the third home video game console produced by Sony Computer Entertainment and the successor to the PlayStation 2 as part of the PlayStation series. The PlayStation 3 competes with Microsoft's Xbox 360 and Nintendo's Wii as part of the seventh generation of video game consoles...
, for a total of about 6.7 native petaFLOPS, (9.1 x86 petaFLOPS) more computing power than the combined efforts of all distributed computing projects under BOINC. A large majority of this performance comes from the GPU and PS3 clients. Folding@home achieves strong scaling across its user base; it gains a near-linear speedup for every additional processor. In 2007, Guinness
Guinness World Records
Guinness World Records, known until 2000 as The Guinness Book of Records , is a reference book published annually, containing a collection of world records, both human achievements and the extremes of the natural world...
recognized Folding@home as the most powerful distributed computing cluster in the world. This large and powerful network allows FAH to do work not possible any other way, including through the use of supercomputers, which are typically expensive to operate and often shared.
Folding@home gained popularity early in its history. In March 2002, Google
Google
Google Inc. is an American multinational public corporation invested in Internet search, cloud computing, and advertising technologies. Google hosts and develops a number of Internet-based services and products, and generates profit primarily from advertising through its AdWords program...
co-founder Sergey Brin
Sergey Brin
Sergey Mikhaylovich Brin is a Russian-born American computer scientist and internet entrepreneur who, with Larry Page, co-founded Google, one of the largest internet companies. , his personal wealth is estimated to be $16.7 billion....
launched Google Compute as add-on for the Google Toolbar
Google Toolbar
Google Toolbar is an Internet browser toolbar only available for Internet Explorer and Firefox .-Google Toolbar 1.0 December 11, 2000:New features:*Direct access to the Google search functionality from any web page*Web Site search...
. Although limited in functionality and scope, it increased Folding@home's participation from 10,000 up to about 30,000 active CPUs. The program ended in October 2005 in favor of the Pande lab's official clients, and is no longer available for the Toolbar. Folding@home also gained participants from Genome@home
Genome@home
Genome@home was a distributed computing project run by Stefan Larson of Stanford University, and a sister project to Folding@home. Its goal was protein design and its applications, which had implications in many fields including medicine...
, another distributed computing project from the Pande lab and a sister project to Folding@home. The goal of Genome@home was protein design
Protein design
Protein design is the design of new protein molecules, either from scratch or by making calculated variations on a known structure. The use of rational design techniques for proteins is a major aspect of protein engineering....
and its applications, and the project was officially concluded in March 2004. Following its completion, users were asked to donate to Folding@home instead.
PetaFLOPS milestones
| Native petaFLOPS threshold | Date crossed | Fastest Supercomputer Supercomputer A supercomputer is a computer at the frontline of current processing capacity, particularly speed of calculation.Supercomputers are used for highly calculation-intensive tasks such as problems including quantum physics, weather forecasting, climate research, molecular modeling A supercomputer is a... at Date Crossed Note 1 |
|---|---|---|
| 1.0 | September 16, 2007 | 0.2806 petaFLOP BlueGene/L |
| 2.0 | May 7, 2008 | 0.4782 petaFLOP BlueGene/L |
| 3.0 | August 20, 2008 | 1.042 petaFLOP Roadrunner |
| 4.0 | September 28, 2008 | 1.042 petaFLOP Roadrunner |
| 5.0 | February 18, 2009 | 1.105 petaFLOP Roadrunner |
| 6.0 | November 10, 2011 | 8.162 petaFLOP K computer K computer The K computer – named for the Japanese word , which stands for 10 quadrillion – is a supercomputer being produced by Fujitsu at the RIKEN Advanced Institute for Computational Science campus in Kobe, Japan. In June 2011, TOP500 ranked K the world's fastest supercomputer, with a rating... |
On September 16, 2007, the Folding@home project officially attained a sustained performance level higher than one native petaFLOPS
FLOPS
In computing, FLOPS is a measure of a computer's performance, especially in fields of scientific calculations that make heavy use of floating-point calculations, similar to the older, simpler, instructions per second...
, becoming the first computing system of any kind in the world to do so, although it had erroneously almost reached that level in March of that year. On May 7, 2008, the project attained a sustained performance level higher than two native petaFLOPS, followed by the three and four native petaFLOPS milestones on August 20 and September 28, 2008 respectively. Then on February 18, 2009, Folding@home achieved a performance level of just above five native petaFLOPS
FLOPS
In computing, FLOPS is a measure of a computer's performance, especially in fields of scientific calculations that make heavy use of floating-point calculations, similar to the older, simpler, instructions per second...
, thereby becoming the first computing system to surpass that performance, just as it was for the other four milestones. Finally, on November 10, 2011, Folding@home crossed the six native petaFLOP barrier with the equivalent of nearly eight x86 petaFLOPS.
Starting in March 2009, Folding@home began reporting performance in both native and x86 FLOPS. While native FLOPS are a measure of the performance from a given hardware, Folding@home also estimates how many FLOPS the calculation would take on the standard x86 architecture, which is commonly used as a performance reference. For instance, certain complex functions can be performed in one native FLOP on a GPU, but take multiple FLOPS on the standard x86 CPU architecture. Despite using conservative conversions, for the GPU and PS3 clients "x86" FLOPS are consistently much greater than the "Native" FLOPS. By reporting in both native and x86 FLOPS, Folding@home attempts to even out these differences.
Points
Distributed computing projects such as Folding@home are often driven by a sense of collegiate competition to compute the most for the project. Folding@home quantitatively assesses this through a point system. Donors are granted point credit as a measure of their contribution, and these points can foster friendly competition between donors. Points are determined by the performance of each contributor's folding hardware relative to a reference machine, and one or more Work Units from a project are benchmarked on that machine before the project is released. As some simulations are exceptionally demanding on a system, or are of great scientific priority, donors who opt-in and reliably complete these Work Units are non-linearly rewarded additional bonus points. This generates a fair system of equal pay for equal work, and attempts to align credit with the value of the scientific results. Donors can also use a passkey to securely protect their contributions, as they not only allow for the receipt of bonus points, but they also separate a donor from any policy issues arising from another using that username.Users can register their contributions under a team, which register the combined score of all their members. A user can start their own team, or they can join an existing team. They can be used for troubleshooting or recruitment purposes, but can also keep donors motivated. In some cases, a team may have their own community-driven sources of help such as a forum
Internet forum
An Internet forum, or message board, is an online discussion site where people can hold conversations in the form of posted messages. They differ from chat rooms in that messages are at least temporarily archived...
. In addition, rivalries between teams create friendly competition that benefits the folding community, and members can also have intra-team competitions for top spots. However, regardless of username or team affiliation, all contributions go to the same place and have the same scientific value. Rankings and other statistics for both individuals and teams are posted to the Folding@home website, with third party statistics sites also available.
Software
Folding@home software on the user's end consists of three components: a client, work units, and cores.Client
Folding@home is powered by volunteers who have installed a clientClient (computing)
A client is an application or system that accesses a service made available by a server. The server is often on another computer system, in which case the client accesses the service by way of a network....
program
Computer program
A computer program is a sequence of instructions written to perform a specified task with a computer. A computer requires programs to function, typically executing the program's instructions in a central processor. The program has an executable form that the computer can use directly to execute...
on their personal computer
Personal computer
A personal computer is any general-purpose computer whose size, capabilities, and original sales price make it useful for individuals, and which is intended to be operated directly by an end-user with no intervening computer operator...
. The project differs from other distributed computing projects, such as those under BOINC, by offering a variety of clients such as those for multi-core processors, graphics processing unit
Graphics processing unit
A graphics processing unit or GPU is a specialized circuit designed to rapidly manipulate and alter memory in such a way so as to accelerate the building of images in a frame buffer intended for output to a display...
s, and PlayStation 3
PlayStation 3
The is the third home video game console produced by Sony Computer Entertainment and the successor to the PlayStation 2 as part of the PlayStation series. The PlayStation 3 competes with Microsoft's Xbox 360 and Nintendo's Wii as part of the seventh generation of video game consoles...
s, complementing the standard client designed for uniprocessor
Uniprocessor
A uniprocessor system is a computer system with a single central processing unit. As more and more computers employ multiprocessing architectures, such as SMP and MPP, the term is used to refer to systems that still have only one CPU. Most desktop computers are now shipped with multiprocessing...
systems. While the former clients use significantly more system resources, they also have the capability of completing an overall simulation very quickly, (in a few weeks or months rather than years) which is of major scientific value. Folding@home is the first project to fully utilize GPUs, PS3s, or multi-core processors for distributed computing. As its software is custom-tailored to each hardware architecture
Hardware architecture
In engineering, hardware architecture refers to the identification of a system's physical components and their interrelationships. This description, often called a hardware design model, allows hardware designers to understand how their components fit into a system architecture and provides...
, Folding@home gains the ability to run many different types of calculations, allowing the Pande Group to address questions previously considered impossible to tackle computationally, and make even greater impacts on knowledge of protein misfolding and its related diseases.
Each client is the software with which the user interacts, and manages the other software components behind the scenes. Through the client, the user may pause the folding process, open an event log, check the work progress, or view personal statistics. These clients run continuously in the background
Background (computer software)
A background process is a computer process that runs "behind the scenes" and without user intervention. Typical tasks for these processes include logging, system monitoring, scheduling, and user notification....
, using otherwise unused processing power. These clients are designed to run FAH's calculations at an extremely low priority, and will back off to allow other computer programs to have more processing power. Although modern computer chips are designed to be able to operate continuously without degrading, if users wish to reduce power consumption or heat production, the maximum percentage of CPU power used can be adjusted if desired. If interrupted by a computer shutdown or other means, the client will resume work at almost the same point at startup. For users with machines with multiple processor units, multiple clients may be installed on one machine, and users may be credited by clients on multiple machines.
For security and scientific integrity reasons, the Pande lab does not publicly release the source code
Source code
In computer science, source code is text written using the format and syntax of the programming language that it is being written in. Such a language is specially designed to facilitate the work of computer programmers, who specify the actions to be performed by a computer mostly by writing source...
of the clients. Significant work goes into minimizing security issues in all of Folding@home's software. For example, clients can be downloaded only from the official Folding@home website or its commercial partners. It will upload and download data only from Stanford's Folding@home data servers, (over port
Computer port (software)
In computer programming, port has a wide range of meanings.A software port is a virtual/logical data connection that can be used by programs to exchange data directly, instead of going through a file or other temporary storage location...
8080, with 80 as an alternative) and will only interact with FAH computer files. Moreover, it does not normally need computer administrative privileges, so from a security standpoint it behaves similar to but is even more secure than a web browser
Web browser
A web browser is a software application for retrieving, presenting, and traversing information resources on the World Wide Web. An information resource is identified by a Uniform Resource Identifier and may be a web page, image, video, or other piece of content...
.
Folding@home's first client was a screensaver
Screensaver
A screensaver is a type of computer program initially designed to prevent phosphor burn-in on CRT and plasma computer monitors by blanking the screen or filling it with moving images or patterns when the computer is not in use...
, which would run Folding@home while the computer was not otherwise in use. Later, the Pande lab tested clients on the open source BOINC framework; however, this approach became unworkable and was abandoned in June 2006. Both BOINC and Folding@home clients fell short, for neither client type had enough ability to be compatible with the other. BOINC lacked many features that FAH needed, and FAH lacked features that BOINC needed.
Graphics processing units
GPUs are computer chips used to accelerate 3D graphics, which are most commonly found in video games. GPUs have the capability to significantly out-perform CPUs in terms of Floating Point OPerations, (FLOPFlop
- Terms :*Flop, a box office bomb in the entertainment world*Flop, as verb or noun, referring to flophouse, cheap rooms in a transients' hotel*Flop , a poker term describing the first three cards dealt to the board...
s) at the cost of lower generality. For this reason, high-performance computing is increasingly utilizing the GPU specialized hardware. In the first large-scale test of GPU scientific reliability, the Pande lab found that although GPUs lack built-in memory error detection and correction
Error detection and correction
In information theory and coding theory with applications in computer science and telecommunication, error detection and correction or error control are techniques that enable reliable delivery of digital data over unreliable communication channels...
, reliable scientific computation can be performed on consumer-grade hardware, as long as sufficient measures are taken (such as Folding@home's built-in error detection) to ensure data integrity.
Despite their potential benefits, scientific computing on GPUs has previously remained inefficient and difficult. The Pande lab has been able to write OpenMM, an open source
Open source
The term open source describes practices in production and development that promote access to the end product's source materials. Some consider open source a philosophy, others consider it a pragmatic methodology...
molecular dynamics library optimized to take full advantage of the GPU architecture and gain large speed increases over conventional single-CPU implementations. As an abstraction layer
Abstraction layer
An abstraction layer is a way of hiding the implementation details of a particular set of functionality...
, OpenMM allows molecular dynamics simulations to be efficiently run across a variety of computer architectures and platforms, something previously problematic in scientific software development. GPUs remain the most powerful platform available in terms of FLOPS
FLOPS
In computing, FLOPS is a measure of a computer's performance, especially in fields of scientific calculations that make heavy use of floating-point calculations, similar to the older, simpler, instructions per second...
; as of September 23, 2011, GPU clients account for 71% of the entire project's FLOP throughput.
The first generation of Folding@home's Windows
Microsoft Windows
Microsoft Windows is a series of operating systems produced by Microsoft.Microsoft introduced an operating environment named Windows on November 20, 1985 as an add-on to MS-DOS in response to the growing interest in graphical user interfaces . Microsoft Windows came to dominate the world's personal...
GPU client (GPU1) was released to the public on October 2, 2006, and delivered a 20-30X speedup for certain calculations over its CPU-based Gromacs counterparts. It was the first time GPUs had been used for either distributed computing or major molecular dynamics calculations. The Pande lab learned much about the development of GPGPU
GPGPU
General-purpose computing on graphics processing units is the technique of using a GPU, which typically handles computation only for computer graphics, to perform computation in applications traditionally handled by the CPU...
software, but citing a need to improve scientific accuracies over DirectX
DirectX
Microsoft DirectX is a collection of application programming interfaces for handling tasks related to multimedia, especially game programming and video, on Microsoft platforms. Originally, the names of these APIs all began with Direct, such as Direct3D, DirectDraw, DirectMusic, DirectPlay,...
, it was succeeded by GPU2, the second generation successor of the client on April 10, 2008. Several months later, GPU1 was officially retired on June 6. Compared to GPU1, GPU2 was more scientifically reliable and productive, ran on ATI
Ati
As a word, Ati may refer to:* Ati, a town in Chad* Ati, a Negrito ethnic group in the Philippines* Ati-Atihan Festival, an annual celebration held in the Philippines* Ati, a queen of the fabled Land of Punt in Africa...
and CUDA
CUDA
CUDA or Compute Unified Device Architecture is a parallel computing architecture developed by Nvidia. CUDA is the computing engine in Nvidia graphics processing units that is accessible to software developers through variants of industry standard programming languages...
-enabled Nvidia
NVIDIA
Nvidia is an American global technology company based in Santa Clara, California. Nvidia is best known for its graphics processors . Nvidia and chief rival AMD Graphics Techonologies have dominated the high performance GPU market, pushing other manufacturers to smaller, niche roles...
GPUs, and supported more advanced algorithms, larger proteins, and real-time visualization of the protein simulation. Following this, the third generation of Folding@home's GPU client (GPU3) was released on May 25, 2010. While backwards compatible to GPU2, GPU3 is more stable and efficient, has additional scientific capabilities, and uses the Pande lab's OpenMM library on top of an OpenCL
OpenCL
OpenCL is a framework for writing programs that execute across heterogeneous platforms consisting of CPUs, GPUs, and other processors. OpenCL includes a language for writing kernels , plus APIs that are used to define and then control the platforms...
framework. Although it does not natively support the Linux
Linux
Linux is a Unix-like computer operating system assembled under the model of free and open source software development and distribution. The defining component of any Linux system is the Linux kernel, an operating system kernel first released October 5, 1991 by Linus Torvalds...
operating system, it can be run under WINE
Wine (software)
Wine is a free software application that aims to allow computer programs written for Microsoft Windows to run on Unix-like operating systems. Wine also provides a software library, known as Winelib, against which developers can compile Windows applications to help port them to Unix-like...
for donors with Nvidia graphics cards.
PlayStation 3

PlayStation 3
The is the third home video game console produced by Sony Computer Entertainment and the successor to the PlayStation 2 as part of the PlayStation series. The PlayStation 3 competes with Microsoft's Xbox 360 and Nintendo's Wii as part of the seventh generation of video game consoles...
s, to achieve performance previously only possible on supercomputers. Unlike Microsoft's Xbox
Xbox
The Xbox is a sixth-generation video game console manufactured by Microsoft. It was released on November 15, 2001 in North America, February 22, 2002 in Japan, and March 14, 2002 in Australia and Europe and is the predecessor to the Xbox 360. It was Microsoft's first foray into the gaming console...
, the PS3 is well suited for Folding@home simulations. At the time of its inception and for certain calculations, its main streaming processor
Stream processing
Stream processing is a computer programming paradigm, related to SIMD , that allows some applications to more easily exploit a limited form of parallel processing...
Cell processor delivered a 20x speed increase over PCs, allowing the Pande lab to address problems previously considered impossible to tackle computationally. This high speed and efficiency introduced other opportunities for worthwhile
Amdahl's law
Amdahl's law, also known as Amdahl's argument, is named after computer architect Gene Amdahl, and is used to find the maximum expected improvement to an overall system when only part of the system is improved...
optimizations, and radically changed the tradeoff between computational efficiency and overall accuracy, allowing for the utilization of more complex molecular models at little extra computational cost. These capabilities allow for greater insights into disease research.
The PS3 client was originally a standalone application, but since September 18, 2008 is a channel of Life with PlayStation
Life with PlayStation
Life with PlayStation is an online multimedia application for the PlayStation 3 video game console on the PlayStation Network. The application has four channels, all of which revolve around a virtual globe that displays information according to the channel...
, developed in a collaborative effort between Sony
Sony
, commonly referred to as Sony, is a Japanese multinational conglomerate corporation headquartered in Minato, Tokyo, Japan and the world's fifth largest media conglomerate measured by revenues....
and the Pande lab. It takes the middle path between a CPU's flexibility and a GPU's speed, performing a limited set of calculations rapidly while still retaining adaptable. However, unlike CPUs and GPUs, donors cannot perform other activities on their PS3 while running Folding@home. Instead, the Pande lab has specifically designed its Work Units to take approximately eight hours so that they can be completed overnight. The PS3's uniform console environment makes support
Technical support
Technical support or tech support refers to a range of services by which enterprises provide assistance to users of technology products such as mobile phones, televisions, computers, software products or other electronic or mechanical goods...
easier, as well as making Folding@home user friendly
User Friendly
User Friendly is a discontinued daily webcomic about the staff of a small, fictional Internet service provider, Columbia Internet. The strip's humor tends to be centered around technology jokes and geek humour....
. The PS3 also has the ability to stream data quickly to its GPU, allowing for real-time atomic detail visualizations of the protein dynamics.
Multi-core processing client
The Symmetric MultiProcessingSymmetric multiprocessing
In computing, symmetric multiprocessing involves a multiprocessor computer hardware architecture where two or more identical processors are connected to a single shared main memory and are controlled by a single OS instance. Most common multiprocessor systems today use an SMP architecture...
(SMP) client fulfills two purposes: it takes advantage of the high-performance capabilities of recent multiprocessor
Multi-core (computing)
A multi-core processor is a single computing component with two or more independent actual processors , which are the units that read and execute program instructions...
systems, and it helps develop a simulation architecture that will become one of the dominant FAH computing paradigms as multi-core chips become an industry standard over the next several years. The SMP client is capable of delivering over a 4x calculation speedup over the standard uniprocessor clients.
Folding@home's SMP core handles multi-core CPUs very different from other distributed computing projects, including those under BOINC. Instead of simply doing multiple Work Units simultaneously, single WUs are completed much faster across the multiple CPU cores. This cuts down on the traditional difficulties of scaling a large simulation to many processors. As such, this approach is very scientifically valuable. Some of the Pande lab publications would not have been possible without the SMP client.
On November 13, 2006, first generation SMP Folding@home clients for x86 Microsoft Windows
Microsoft Windows
Microsoft Windows is a series of operating systems produced by Microsoft.Microsoft introduced an operating environment named Windows on November 20, 1985 as an add-on to MS-DOS in response to the growing interest in graphical user interfaces . Microsoft Windows came to dominate the world's personal...
, x86-64
X86-64
x86-64 is an extension of the x86 instruction set. It supports vastly larger virtual and physical address spaces than are possible on x86, thereby allowing programmers to conveniently work with much larger data sets. x86-64 also provides 64-bit general purpose registers and numerous other...
Linux
Linux
Linux is a Unix-like computer operating system assembled under the model of free and open source software development and distribution. The defining component of any Linux system is the Linux kernel, an operating system kernel first released October 5, 1991 by Linus Torvalds...
, and x86 Mac OS X
Mac OS X
Mac OS X is a series of Unix-based operating systems and graphical user interfaces developed, marketed, and sold by Apple Inc. Since 2002, has been included with all new Macintosh computer systems...
were released. These clients used Message Passing Interface
Message Passing Interface
Message Passing Interface is a standardized and portable message-passing system designed by a group of researchers from academia and industry to function on a wide variety of parallel computers...
(MPI) protocols on the localhost
Localhost
In computer networking, localhost is the standard hostname given to the address of the loopback network interface. The name is also a reserved top-level domain name In computer networking, localhost (meaning this computer) is the standard hostname given to the address of the loopback network...
, as at the time the Gromacs cores were not designed to be used with multiple thread
Thread (computer science)
In computer science, a thread of execution is the smallest unit of processing that can be scheduled by an operating system. The implementation of threads and processes differs from one operating system to another, but in most cases, a thread is contained inside a process...
s. This made Folding@home the first to use MPI for distributed computing software, as it had previously been reserved only for supercomputers. The MPI-based clients worked well in Unix
Unix
Unix is a multitasking, multi-user computer operating system originally developed in 1969 by a group of AT&T employees at Bell Labs, including Ken Thompson, Dennis Ritchie, Brian Kernighan, Douglas McIlroy, and Joe Ossanna...
-based operating systems such as Linux
Linux
Linux is a Unix-like computer operating system assembled under the model of free and open source software development and distribution. The defining component of any Linux system is the Linux kernel, an operating system kernel first released October 5, 1991 by Linus Torvalds...
and Mac's OS-X
OS-X
OS-X is a priority-based pre-emptive multitasking real-time embedded operating system designed for embedded Zilog Z80 systems with bank-switched memory...
, but was particularly troublesome in Windows. Despite these difficulties, SMP1 generated significant results that would have been impossible otherwise and which represented a landmark in the simulation of protein folding.
The second generation of the SMP client was released as an open beta on January 24, 2010, and subsequently replaced SMP1. The SMP2 client exchanges the complex MPI for thread
Thread (computer science)
In computer science, a thread of execution is the smallest unit of processing that can be scheduled by an operating system. The implementation of threads and processes differs from one operating system to another, but in most cases, a thread is contained inside a process...
s, which removes much of the overhead of keeping the cores synchronized. The SMP2 client also supports a bonus points system, which non-linearly rewards additional points to donors for quick WU returns and for contributing to next-generation capabilities. Donors who run the SMP2 client receive these extra points if they use a passkey and maintained an 80% successful return of Work Units.
SMP2 also supports extra-large Work Units for users with powerful eight-core CPUs or better. While these WUs consume even more RAM
Ram
-Animals:*Ram, an uncastrated male sheep*Ram cichlid, a species of freshwater fish endemic to Colombia and Venezuela-Military:*Battering ram*Ramming, a military tactic in which one vehicle runs into another...
and have more network usage than regular SMP WUs, users who run these are rewarded with a 20% increase over SMP2's bonus point system. These powerful computers allow for simulations to be performed on Folding@home that had previously required the use of supercomputing clusters. There is a great scientific need to run these simulations out to long timescales as quickly as possible, so the additional bonus points also serves as an incentive for rapid completions of Work Units. This allows the Pande lab to perform studies of larger molecular systems that would not have been possible anywhere else on Folding@home.
V7
The v7 client is the seventh and latest generation of the Folding@home software, currently under development, but available for open beta testing. V7 is a complete rewrite and unification of the previous clients for Microsoft WindowsMicrosoft Windows
Microsoft Windows is a series of operating systems produced by Microsoft.Microsoft introduced an operating environment named Windows on November 20, 1985 as an add-on to MS-DOS in response to the growing interest in graphical user interfaces . Microsoft Windows came to dominate the world's personal...
, Mac OS X
Mac OS X
Mac OS X is a series of Unix-based operating systems and graphical user interfaces developed, marketed, and sold by Apple Inc. Since 2002, has been included with all new Macintosh computer systems...
and Linux
Linux
Linux is a Unix-like computer operating system assembled under the model of free and open source software development and distribution. The defining component of any Linux system is the Linux kernel, an operating system kernel first released October 5, 1991 by Linus Torvalds...
operating systems. Following its predecessors, v7 runs Folding@home in the background at very low priority, which allows other applications to use CPU resources as they need. The v7 client is designed to make the installation, start-up, and operation user-friendly for novices, as well as offer greater scientific flexibility than previous clients. It is the Pande lab's goal to make v7 the recommended client by January 2012 at the latest, and versions of v7 will be frequently released until then.
V7 consists of several elements. The user interacts with v7's GUI
Gui
Gui or guee is a generic term to refer to grilled dishes in Korean cuisine. These most commonly have meat or fish as their primary ingredient, but may in some cases also comprise grilled vegetables or other vegetarian ingredients. The term derives from the verb, "gupda" in Korean, which literally...
, known as FAHControl. It has Novice, Advanced, and Expert user interface modes, and has the ability to monitor, configure, and control many remote folding clients from a single computer. FAHControl can monitor and direct FAHClient, which runs behind the scenes and in turn manages each FAHSlot (or "slot"). These slots act as replacements for the previously distinct FAH clients, as they may be of Uniprocessor, SMP, or GPU type. Each slot also contains a core and data associated with it, and can download, process, and upload Work Units independently. The FAHViewer function, modeled after the PS3 viewer, displays a real-time 3D rendering, if available, of the protein currently being processed.
Work Units
The Work Unit (WU) is the protein data that the client is being asked to process. Each WU is identified for its respective protein Project, Run (conformation), Clone (atomic trajectory), and Generation (time steps in the trajectory/simulation). The client connects to Folding@home serverServer (computing)
In the context of client-server architecture, a server is a computer program running to serve the requests of other programs, the "clients". Thus, the "server" performs some computational task on behalf of "clients"...
s to retrieve a Work Unit, processes it, and returns it upon completion. During transfer, all Work Units are validated through the use of 2048-bit digital signature
Digital signature
A digital signature or digital signature scheme is a mathematical scheme for demonstrating the authenticity of a digital message or document. A valid digital signature gives a recipient reason to believe that the message was created by a known sender, and that it was not altered in transit...
s. These WUs have associated deadlines and credit (point) value. If this deadline is exceeded, the user may not get credit and the unit will be reissued to someone else. As protein folding is serial in nature and each WU is generated from its predecessor, this allows the overall simulation process to proceed normally if a WU is not returned after a certain period of time. Due to these deadlines, the minimum system requirements for Folding@home is a Pentium 3 450 MHz CPU with SSE
SSE
-Computing:*Server-sent events, a technology to push content to web clients*Simple Sharing Extensions, a specification that extends RSS from unidirectional to bidirectional information flows*SPARQL Syntax Expressions*Microsoft SQL Server Express Edition...
or newer. However, Work Units for high performance clients have a much shorter deadline than those for the uniprocessor client, as a major part of the scientific benefit is dependent on rapidly completing simulations.
Before public release, Work Units go through several Quality Assurance
Quality Assurance
Quality assurance, or QA for short, is the systematic monitoring and evaluation of the various aspects of a project, service or facility to maximize the probability that minimum standards of quality are being attained by the production process...
steps to keep problematic WUs from becoming fully available. But unlike particular BOINC projects such as SETI@home
SETI@home
SETI@home is an Internet-based public volunteer computing project employing the BOINC software platform, hosted by the Space Sciences Laboratory, at the University of California, Berkeley, in the United States. SETI is an acronym for the Search for Extra-Terrestrial Intelligence...
, Folding@home's Work Units are normally processed only once, except in the rare event that errors occur during processing of a WU. If this occurs for three different donors, it is automatically pulled from distribution. Topics in the Folding@home forum can be used to differentiate between problematic hardware and an actual bad Work Unit.
Work Units are very much tied to the Pande lab's simulation Markov state models
Hidden Markov model
A hidden Markov model is a statistical Markov model in which the system being modeled is assumed to be a Markov process with unobserved states. An HMM can be considered as the simplest dynamic Bayesian network. The mathematics behind the HMM was developed by L. E...
, which allow for extensive parallelization
Parallel computing
Parallel computing is a form of computation in which many calculations are carried out simultaneously, operating on the principle that large problems can often be divided into smaller ones, which are then solved concurrently . There are several different forms of parallel computing: bit-level,...
of very long simulation processes which otherwise seem intrinsically serial. During the folding process, proteins spend much of their time "waiting" in various states, before quickly transitioning to the next configuration. This allows for the unique possibility to simulate only a small fraction of the overall folding timescale, leading to significant speedups. The Pande lab achieves this by first dividing the protein's possible dynamics into a series of related conformation states, and creates WUs to calculate the rates of transition between these states. When the completed WUs are gathered, the Pande lab then runs sophisticated Bayesian Machine Learning
Machine learning
Machine learning, a branch of artificial intelligence, is a scientific discipline concerned with the design and development of algorithms that allow computers to evolve behaviors based on empirical data, such as from sensor data or databases...
algorithms which calculate which states are reasonable and the rates between them. This also averages
Ensemble average
In statistical mechanics, the ensemble average is defined as the mean of a quantity that is a function of the micro-state of a system , according to the distribution of the system on its micro-states in this ensemble....
the molecular simulation ensembles, which is important so that direct, meaningful comparison between in silico simulations and in vitro experiments can be made. This system is successful even at the millisecond timescale, compares well to traditional methods and experimental results, and allows for previously intractable problems to be within reach.
Cores
Specialized scientific computer programs, referred to as "cores," perform the calculations on the Work Unit behind the scenes. Folding@home's cores modified and optimized versions of molecular dynamicsMolecular dynamics
Molecular dynamics is a computer simulation of physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a period of time, giving a view of the motion of the atoms...
programs, including GROMACS, AMBER
AMBER
AMBER is a family of force fields for molecular dynamics of biomolecules originally developed by the late Peter Kollman's group at the University of California, San Francisco. AMBER is also the name for the molecular dynamics software package that simulates these force fields...
, TINKER
TINKER
TINKER is a computer software application for molecular dynamics simulation with a complete and general package for molecular mechanics and molecular dynamics, with some special features for biopolymers...
, CPMD
CPMD
The Car–Parrinello Molecular Dynamics,, also known as CPMD, is a method to follow the classical motion of point-like atomic nuclei in time, i.e. for performing molecular dynamics , while at the same time solving ab-initio and efficiently the quantum mechanical motion of electrons...
, SHARPEN, ProtoMol, BrookGPU
BrookGPU
BrookGPU is the Stanford University graphics group's compiler and runtime implementation of the Brook stream programming language for using modern graphics hardware for non-graphical, general purpose computations...
and Desmond
Desmond (software)
Desmond is a software package developed at D. E. Shaw Research to perform high-speed molecular dynamics simulations of biological systems on conventional computer clusters. The code uses novel parallel algorithms and numerical techniques to achieve high performance on platforms containing a large...
. Some of these cores perform explicit atom-by-atom molecular dynamics
Molecular dynamics
Molecular dynamics is a computer simulation of physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a period of time, giving a view of the motion of the atoms...
calculations, while others perform implicit solvation
Implicit solvation
Implicit solvation is a method of representing solvent as a continuous medium instead of individual “explicit” solvent molecules most often used in molecular dynamics simulations and in other applications of molecular mechanics...
methods, which treat atoms as a mathematical continuum. These cores are open-source software
Open-source software
Open-source software is computer software that is available in source code form: the source code and certain other rights normally reserved for copyright holders are provided under a software license that permits users to study, change, improve and at times also to distribute the software.Open...
or are under similar licenses, and are verified during download by 2048-bit digital signatures. While the same core can be used by various versions of the client, separating the core from the client enables the scientific methods to be updated automatically as needed without a client update.
Comparison to other molecular systems
Rosetta@homeRosetta@home
Rosetta@home is a distributed computing project for protein structure prediction on the Berkeley Open Infrastructure for Network Computing platform, run by the Baker laboratory at the University of Washington...
is a distributed computing project aimed at protein structure prediction
Protein structure prediction
Protein structure prediction is the prediction of the three-dimensional structure of a protein from its amino acid sequence — that is, the prediction of its secondary, tertiary, and quaternary structure from its primary structure. Structure prediction is fundamentally different from the inverse...
and is one of the most successful approaches to this problem. Folding@home and Rosetta@home address very different molecular questions. Although Rosetta@home does not provide information into how proteins fold, it does predict the protein's most likely final structure, which in some cases is used as a basis for Folding@home's projects. Rosetta's predictions can help FAH simulate the folding of larger proteins more efficiently. Folding@home can also verify Rosetta@home's results and find additional atomistic details of the protein's kinetics and folding pathway, which is intrinsically much more difficult. Folding@home's accurate simulations have also suggesting important novel implications into the fields of protein folding, structure prediction, and certain folding experiments, and have shown that Rosetta's structure prediction may benefit from thermodynamic sampling aspects of protein folding mechanisms.
Folding@home also compares well to Anton
Anton (computer)
Anton is a massively parallel supercomputer designed and built by D. E. Shaw Research in New York. It is a special-purpose system for molecular dynamics simulations of proteins and other biological macromolecules...
, a powerful supercomputer which uses specialized hardware to produce a small number of ultra-long molecular trajectories. It is unique in this ability, and like Folding@home, has also improved particular long-held theories of protein folding. Its longer simulations, while computationally expensive, contain more phase space
Phase space
In mathematics and physics, a phase space, introduced by Willard Gibbs in 1901, is a space in which all possible states of a system are represented, with each possible state of the system corresponding to one unique point in the phase space...
than any one of Folding@home's many shorter trajectories, which allows Anton to perform a thorough exploration of the required space. As of October 2011, Anton and FAH are the two most powerful molecular dynamics systems, and Anton has also run simulations out to the millisecond range. In 2011, the Pande lab built a Markov state model from one of Anton's simulations. It demonstrated that there was little difference between MSMs built from Anton's fewer long trajectories and one assembled from Folding@home's many shorter trajectories. Their analysis also showed that Folding@home's Markov state models significantly improve the analysis of these longer simulations, such as revealing additional relevant folding pathways and information into how the protein carries out its biological function. Folding@home is running further analysis on one of Anton's simulations to better determine how its approaches compare to Anton's methods. It is probable that a combination of Anton's and FAH's simulation methods would be very beneficial, and Pande looks forward to see how Anton and FAH can be used together.
See also
- Storage@homeStorage@homeStorage@home is a distributed storage infrastructure designed to store massive amounts of scientific data across a large host of volunteer machines.The project was developed by some of the Folding@home team at Stanford University.- Function :...
- List of distributed computing projects
- Software for molecular modeling
- Molecular modeling on GPUMolecular modeling on GPUMolecular modeling on GPU is the technique of using a graphics processing unit for molecular simulations.In 2007, NVIDIA introduced video cards that could be used not only to show graphics but also for scientific calculations. These cards include many arithmetic units working in parallel...
s - Rosetta@homeRosetta@homeRosetta@home is a distributed computing project for protein structure prediction on the Berkeley Open Infrastructure for Network Computing platform, run by the Baker laboratory at the University of Washington...
- Blue GeneBlue GeneBlue Gene is a computer architecture project to produce several supercomputers, designed to reach operating speeds in the PFLOPS range, and currently reaching sustained speeds of nearly 500 TFLOPS . It is a cooperative project among IBM Blue Gene is a computer architecture project to produce...
- Molecular dynamicsMolecular dynamicsMolecular dynamics is a computer simulation of physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a period of time, giving a view of the motion of the atoms...
- Computational biologyComputational biologyComputational biology involves the development and application of data-analytical and theoretical methods, mathematical modeling and computational simulation techniques to the study of biological, behavioral, and social systems...
External links
- Folding@home homepage
- Folding@home official blog
- Folding@home forum
- Folding@home statistics
- List of publications from Folding@home's results
- "Futures In Biotech" episode (for newcomers)
- 2009 Interview with Vijay Pande about Folding@home Project
- Video of record-breaking 1.5ms protein fold
- Pande lab's OpenMM molecular dynamics library
- Simple multimedia presentation about Folding@home
- Folding@home Wiki
- Wikipedia Team

