
Genome size
Encyclopedia
Genome size is the total amount of DNA
contained within one copy of a single genome
. It is typically measured in terms of mass
in picograms (trillionths (10−12) of a gram
, abbreviated pg) or less frequently in Daltons or as the total number of nucleotide
base pairs typically in megabases (millions of base pairs, abbreviated Mb or Mbp). One picogram equals 978 megabases. In diploid organisms, genome size is used interchangeably with the term C-value
.
An organism's complexity is not directly proportional to its genome size; some single cell organisms have much more DNA than humans (see Junk DNA and C-value enigma
).
DNA content does, in fact, reflect genome size". In this context, "genome size" was being used in the sense of genotype
to mean the number of genes
. In a paper submitted only two months later (in February 1969), Wolf et al. (1969) used the term "genome size" throughout and in its present usage; therefore these authors should probably be credited with originating the term in its modern sense. By the early 1970s, "genome size" was in common usage with its present definition, probably as a result of its inclusion in Susumu Ohno
's influential book Evolution by Gene Duplication, published in 1970.
measurements of Feulgen-stained nuclei (previously using specialized densitometers, now more commonly using computerized image analysis
) or flow cytometry
. In prokaryotes, pulsed field gel electrophoresis
and complete genome sequencing are the predominant methods of genome size determination. Nuclear genome sizes are well known to vary enormously among eukaryotic species. In animals they range more than 3,300-fold, and in land plants they differ by a factor of about 1,000. Protist
genomes have been reported to vary more than 300,000-fold in size, but the high end of this range (Amoeba
) has been called into question. In eukaryotes (but not prokaryotes), variation in genome size is not proportional to the number of genes
, an observation that was deemed wholly counterintuitive before the discovery of non-coding DNA and which became known as the C-value paradox as a result. However, although there is no longer any paradox
ical aspect to the discrepancy between genome size and gene number, this term remains in common usage. For reasons of conceptual clarification, the various puzzles that remain with regard to genome size variation instead have been suggested by one author to more accurately comprise a puzzle or an enigma (the C-value enigma
). Genome size correlates with a range of features at the cell
and organism levels, including cell size, cell division
rate, and, depending on the taxon
, body size, metabolic rate, developmental rate, organ
complexity, geographical distribution, and/or extinction
risk (for recent reviews, see Bennett and Leitch 2005; Gregory 2005). Based on completely sequenced genome data currently (as of April 2009) available, log-transformed gene number forms a linear correlation with log-transformed genome size in bacteria, archea, viruses, and organelles combined whereas a nonlinear (semi-natural log) correlation in eukaryotes (Hou and Lin 2009 ). The nonlinear correlation for eukaryotes, although claim of its existence contrasts the previous view that no correlation exists for this group of organisms, reflects disproportinately fast increasing noncoding DNA in increasingly large eukaryotic genomes. Although sequenced genome data are practically biased toward small genomes, which may compromise the accuracy of the empirically derived correlation, and the ultimate proof of the correlation remains to be obtained by sequencing some of the largest eukaryotic genomes, current data do not seem to rule out a correlation.
shrinks relative to its ancestor. Genomes fluctuate in size regularly, especially in Bacteria
, but in some situations a genome has drastically lost content during some period.
The most evolution
ary significant cases of genome reduction may be the eukaryotic organelle
s that are derived from bacteria: the mitochondrion
and plastid
. These organelles are descended from endosymbiont
s, which can only survive within the host cell and which the host cell likewise needs for survival. Many mitochondria have less than 20 genes in their entire genome, whereas a free-living bacterium generally has at least 1000 genes. Many genes have been transferred to the host nucleus
, while others have simply been lost and their function replaced by host processes.
Other bacteria have become endosymbionts or obligate intracellular pathogen
s and experienced extensive genome reduction as a result. This process seems to be dominated by genetic drift
resulting from small population
size, low recombination
rates, and high mutation
rates, as opposed to selection
for smaller genomes.
Some free-living marine bacterioplanktons also shows signs of genome reduction, which are hypothesized to be driven by natural selection.
environment. These species have become a considerable threat to human health, as they are often highly capable of evading human immune systems and manipulating the host environment to acquire nutrients. A common explanation for these keen manipulative abilities is the compact and efficient genomic structure consistently found in obligate endosymbionts. This compact genome structure is the result of massive losses of extraneous DNA - an occurrence that is exclusively associated with the loss of a free-living stage. In fact, as much as 90% of the genetic material can be lost when a species makes the evolutionary transition from a free-living to obligate intracellular lifestyle. Common examples of species with reduced genomes include: Buchnera aphidicola, Rickettsia
prowazekii and Mycobacterium leprae
. One obligate endosymbiont of psyllid, Candidatus Carsonella ruddii
, has the smallest genome currently known among cellular organisms at 160kb. It is important to note, however, that some obligate intracellular species have positive fitness effects on their hosts. (See also mutualists and parasites.)
The reductive evolution model has been proposed as an effort to define the genomic commonalities seen in all obligate endosymbionts. This model illustrates four general features of reduced genomes and obligate intracellular species:
Based on this model, it is clear that endosymbionts face different adaptive challenges than free-living species.

or simply: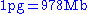
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
contained within one copy of a single genome
Genome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
. It is typically measured in terms of mass
Mass
Mass can be defined as a quantitive measure of the resistance an object has to change in its velocity.In physics, mass commonly refers to any of the following three properties of matter, which have been shown experimentally to be equivalent:...
in picograms (trillionths (10−12) of a gram
Gram
The gram is a metric system unit of mass....
, abbreviated pg) or less frequently in Daltons or as the total number of nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
base pairs typically in megabases (millions of base pairs, abbreviated Mb or Mbp). One picogram equals 978 megabases. In diploid organisms, genome size is used interchangeably with the term C-value
C-value
The term C-value refers to the amount of DNA contained within a haploid nucleus or one half the amount in a diploid somatic cell of a eukaryotic organism, expressed in picograms...
.
An organism's complexity is not directly proportional to its genome size; some single cell organisms have much more DNA than humans (see Junk DNA and C-value enigma
C-value enigma
The C-value enigma or C-value paradox is a term used to describe the complex puzzle surrounding the extensive variation in nuclear genome size among eukaryotic species...
).
Origin of the term
The term "genome size" is often erroneously attributed to Hinegardner, even in discussions dealing specifically with terminology in this area of research (e.g., Greilhuber, 2005). Notably, Hinegardner used the term only once: in the title. The term actually seems to have first appeared in 1968 when Hinegardner wondered, in the last paragraph of his article, whether "cellularCell (biology)
The cell is the basic structural and functional unit of all known living organisms. It is the smallest unit of life that is classified as a living thing, and is often called the building block of life. The Alberts text discusses how the "cellular building blocks" move to shape developing embryos....
DNA content does, in fact, reflect genome size". In this context, "genome size" was being used in the sense of genotype
Genotype
The genotype is the genetic makeup of a cell, an organism, or an individual usually with reference to a specific character under consideration...
to mean the number of genes
Gênes
Gênes is the name of a département of the First French Empire in present Italy, named after the city of Genoa. It was formed in 1805, when Napoleon Bonaparte occupied the Republic of Genoa. Its capital was Genoa, and it was divided in the arrondissements of Genoa, Bobbio, Novi Ligure, Tortona and...
. In a paper submitted only two months later (in February 1969), Wolf et al. (1969) used the term "genome size" throughout and in its present usage; therefore these authors should probably be credited with originating the term in its modern sense. By the early 1970s, "genome size" was in common usage with its present definition, probably as a result of its inclusion in Susumu Ohno
Susumu Ohno
was an Asian American geneticist and evolutionary biologist, and seminal researcher in the field of molecular evolution.- Biography :Susumu Ohno was born of Japanese parents in Seoul, Korea, on February 1, 1928. The second of five children, he was the son of the minister of education of the...
's influential book Evolution by Gene Duplication, published in 1970.
Variation in genome size and gene content
The genome sizes of thousands of eukaryotes have been analyzed over the past 50 years, and these data are available in online databases for animals, plants, and fungi (see external links). Nuclear genome size is typically measured in eukaryotes using either densitometricDensitometry
Densitometry is the quantitative measurement of optical density in light-sensitive materials, such as photographic paper or film, due to exposure to light...
measurements of Feulgen-stained nuclei (previously using specialized densitometers, now more commonly using computerized image analysis
Image analysis
Image analysis is the extraction of meaningful information from images; mainly from digital images by means of digital image processing techniques...
) or flow cytometry
Flow cytometry
Flow cytometry is a technique for counting and examining microscopic particles, such as cells and chromosomes, by suspending them in a stream of fluid and passing them by an electronic detection apparatus. It allows simultaneous multiparametric analysis of the physical and/or chemical...
. In prokaryotes, pulsed field gel electrophoresis
Pulsed field gel electrophoresis
Pulsed field gel electrophoresis is a technique used for the separation of large deoxyribonucleic acid molecules by applying an electric field that periodically changes direction to a gel matrix.-Historical background:...
and complete genome sequencing are the predominant methods of genome size determination. Nuclear genome sizes are well known to vary enormously among eukaryotic species. In animals they range more than 3,300-fold, and in land plants they differ by a factor of about 1,000. Protist
Protist
Protists are a diverse group of eukaryotic microorganisms. Historically, protists were treated as the kingdom Protista, which includes mostly unicellular organisms that do not fit into the other kingdoms, but this group is contested in modern taxonomy...
genomes have been reported to vary more than 300,000-fold in size, but the high end of this range (Amoeba
Amoeba
Amoeba is a genus of Protozoa.History=The amoeba was first discovered by August Johann Rösel von Rosenhof in 1757. Early naturalists referred to Amoeba as the Proteus animalcule after the Greek god Proteus, who could change his shape...
) has been called into question. In eukaryotes (but not prokaryotes), variation in genome size is not proportional to the number of genes
Gênes
Gênes is the name of a département of the First French Empire in present Italy, named after the city of Genoa. It was formed in 1805, when Napoleon Bonaparte occupied the Republic of Genoa. Its capital was Genoa, and it was divided in the arrondissements of Genoa, Bobbio, Novi Ligure, Tortona and...
, an observation that was deemed wholly counterintuitive before the discovery of non-coding DNA and which became known as the C-value paradox as a result. However, although there is no longer any paradox
Paradox
Similar to Circular reasoning, A paradox is a seemingly true statement or group of statements that lead to a contradiction or a situation which seems to defy logic or intuition...
ical aspect to the discrepancy between genome size and gene number, this term remains in common usage. For reasons of conceptual clarification, the various puzzles that remain with regard to genome size variation instead have been suggested by one author to more accurately comprise a puzzle or an enigma (the C-value enigma
C-value enigma
The C-value enigma or C-value paradox is a term used to describe the complex puzzle surrounding the extensive variation in nuclear genome size among eukaryotic species...
). Genome size correlates with a range of features at the cell
Cell (biology)
The cell is the basic structural and functional unit of all known living organisms. It is the smallest unit of life that is classified as a living thing, and is often called the building block of life. The Alberts text discusses how the "cellular building blocks" move to shape developing embryos....
and organism levels, including cell size, cell division
Cell division
Cell division is the process by which a parent cell divides into two or more daughter cells . Cell division is usually a small segment of a larger cell cycle. This type of cell division in eukaryotes is known as mitosis, and leaves the daughter cell capable of dividing again. The corresponding sort...
rate, and, depending on the taxon
Taxon
|thumb|270px|[[African elephants]] form a widely-accepted taxon, the [[genus]] LoxodontaA taxon is a group of organisms, which a taxonomist adjudges to be a unit. Usually a taxon is given a name and a rank, although neither is a requirement...
, body size, metabolic rate, developmental rate, organ
Organ (anatomy)
In biology, an organ is a collection of tissues joined in structural unit to serve a common function. Usually there is a main tissue and sporadic tissues . The main tissue is the one that is unique for the specific organ. For example, main tissue in the heart is the myocardium, while sporadic are...
complexity, geographical distribution, and/or extinction
Extinction
In biology and ecology, extinction is the end of an organism or of a group of organisms , normally a species. The moment of extinction is generally considered to be the death of the last individual of the species, although the capacity to breed and recover may have been lost before this point...
risk (for recent reviews, see Bennett and Leitch 2005; Gregory 2005). Based on completely sequenced genome data currently (as of April 2009) available, log-transformed gene number forms a linear correlation with log-transformed genome size in bacteria, archea, viruses, and organelles combined whereas a nonlinear (semi-natural log) correlation in eukaryotes (Hou and Lin 2009 ). The nonlinear correlation for eukaryotes, although claim of its existence contrasts the previous view that no correlation exists for this group of organisms, reflects disproportinately fast increasing noncoding DNA in increasingly large eukaryotic genomes. Although sequenced genome data are practically biased toward small genomes, which may compromise the accuracy of the empirically derived correlation, and the ultimate proof of the correlation remains to be obtained by sequencing some of the largest eukaryotic genomes, current data do not seem to rule out a correlation.
Genome reduction
Genome reduction, also known as Genome degradation, is the process by which a genomeGenome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
shrinks relative to its ancestor. Genomes fluctuate in size regularly, especially in Bacteria
Bacteria
Bacteria are a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a wide range of shapes, ranging from spheres to rods and spirals...
, but in some situations a genome has drastically lost content during some period.
The most evolution
Evolution
Evolution is any change across successive generations in the heritable characteristics of biological populations. Evolutionary processes give rise to diversity at every level of biological organisation, including species, individual organisms and molecules such as DNA and proteins.Life on Earth...
ary significant cases of genome reduction may be the eukaryotic organelle
Organelle
In cell biology, an organelle is a specialized subunit within a cell that has a specific function, and is usually separately enclosed within its own lipid bilayer....
s that are derived from bacteria: the mitochondrion
Mitochondrion
In cell biology, a mitochondrion is a membrane-enclosed organelle found in most eukaryotic cells. These organelles range from 0.5 to 1.0 micrometers in diameter...
and plastid
Plastid
Plastids are major organelles found in the cells of plants and algae. Plastids are the site of manufacture and storage of important chemical compounds used by the cell...
. These organelles are descended from endosymbiont
Endosymbiont
An endosymbiont is any organism that lives within the body or cells of another organism, i.e. forming an endosymbiosis...
s, which can only survive within the host cell and which the host cell likewise needs for survival. Many mitochondria have less than 20 genes in their entire genome, whereas a free-living bacterium generally has at least 1000 genes. Many genes have been transferred to the host nucleus
Cell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
, while others have simply been lost and their function replaced by host processes.
Other bacteria have become endosymbionts or obligate intracellular pathogen
Pathogen
A pathogen gignomai "I give birth to") or infectious agent — colloquially, a germ — is a microbe or microorganism such as a virus, bacterium, prion, or fungus that causes disease in its animal or plant host...
s and experienced extensive genome reduction as a result. This process seems to be dominated by genetic drift
Genetic drift
Genetic drift or allelic drift is the change in the frequency of a gene variant in a population due to random sampling.The alleles in the offspring are a sample of those in the parents, and chance has a role in determining whether a given individual survives and reproduces...
resulting from small population
Population genetics
Population genetics is the study of allele frequency distribution and change under the influence of the four main evolutionary processes: natural selection, genetic drift, mutation and gene flow. It also takes into account the factors of recombination, population subdivision and population...
size, low recombination
Genetic recombination
Genetic recombination is a process by which a molecule of nucleic acid is broken and then joined to a different one. Recombination can occur between similar molecules of DNA, as in homologous recombination, or dissimilar molecules, as in non-homologous end joining. Recombination is a common method...
rates, and high mutation
Mutation
In molecular biology and genetics, mutations are changes in a genomic sequence: the DNA sequence of a cell's genome or the DNA or RNA sequence of a virus. They can be defined as sudden and spontaneous changes in the cell. Mutations are caused by radiation, viruses, transposons and mutagenic...
rates, as opposed to selection
Natural selection
Natural selection is the nonrandom process by which biologic traits become either more or less common in a population as a function of differential reproduction of their bearers. It is a key mechanism of evolution....
for smaller genomes.
Some free-living marine bacterioplanktons also shows signs of genome reduction, which are hypothesized to be driven by natural selection.
Genome reduction in obligate endosymbiotic species
Obligate endosymbiotic species are characterized by a complete inability to survive external to their hostHost (biology)
In biology, a host is an organism that harbors a parasite, or a mutual or commensal symbiont, typically providing nourishment and shelter. In botany, a host plant is one that supplies food resources and substrate for certain insects or other fauna...
environment. These species have become a considerable threat to human health, as they are often highly capable of evading human immune systems and manipulating the host environment to acquire nutrients. A common explanation for these keen manipulative abilities is the compact and efficient genomic structure consistently found in obligate endosymbionts. This compact genome structure is the result of massive losses of extraneous DNA - an occurrence that is exclusively associated with the loss of a free-living stage. In fact, as much as 90% of the genetic material can be lost when a species makes the evolutionary transition from a free-living to obligate intracellular lifestyle. Common examples of species with reduced genomes include: Buchnera aphidicola, Rickettsia
Rickettsia
Rickettsia is a genus of non-motile, Gram-negative, non-sporeforming, highly pleomorphic bacteria that can present as cocci , rods or thread-like . Being obligate intracellular parasites, the Rickettsia survival depends on entry, growth, and replication within the cytoplasm of eukaryotic host cells...
prowazekii and Mycobacterium leprae
Mycobacterium leprae
Mycobacterium leprae, also known as Hansen’s coccus spirilly, mostly found in warm tropical countries, is a bacterium that causes leprosy . It is an intracellular, pleomorphic, acid-fast bacterium. M. leprae is an aerobic bacillus surrounded by the characteristic waxy coating unique to mycobacteria...
. One obligate endosymbiont of psyllid, Candidatus Carsonella ruddii
Candidatus Carsonella ruddii
Candidatus Carsonella ruddii is an obligate endosymbiotic Gamma Proteobacterium; it has the smallest genome of any characterised bacteria....
, has the smallest genome currently known among cellular organisms at 160kb. It is important to note, however, that some obligate intracellular species have positive fitness effects on their hosts. (See also mutualists and parasites.)
The reductive evolution model has been proposed as an effort to define the genomic commonalities seen in all obligate endosymbionts. This model illustrates four general features of reduced genomes and obligate intracellular species:
- ‘genome streamlining’ resulting from relaxed selection on genes that are superfluous in the intracellular environment;
- a bias towards deletions (rather than insertions), which heavily affects genes that have been disrupted by accumulation of mutations (pseudogenes);
- very little or no capability for acquiring new DNA; and
- considerable reduction of effective population sizeEffective population sizeIn population genetics, the concept of effective population size Ne was introduced by the American geneticist Sewall Wright, who wrote two landmark papers on it...
in endosymbiotic populations, particularly in species that rely on vertical transmission.
Based on this model, it is clear that endosymbionts face different adaptive challenges than free-living species.
Conversion from picograms (pg) to base pairs (bp)

or simply:
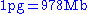
See also
- Comparison of different genome sizes
- Animal Genome Size DatabaseAnimal Genome Size DatabaseThe Animal Genome Size Database is a comprehensive catalogue of published genome size estimates for vertebrate and invertebrate animals. It was created in 2001 by Dr. T. Ryan Gregory of the University of Guelph in Canada. As of September 2005, the database contains data for over 4,000 species of...
- Cell nucleusCell nucleusIn cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
- Comparative genomicsComparative genomicsComparative genomics is the study of the relationship of genome structure and function across different biological species or strains. Comparative genomics is an attempt to take advantage of the information provided by the signatures of selection to understand the function and evolutionary...
- C-valueC-valueThe term C-value refers to the amount of DNA contained within a haploid nucleus or one half the amount in a diploid somatic cell of a eukaryotic organism, expressed in picograms...
- C-value enigmaC-value enigmaThe C-value enigma or C-value paradox is a term used to describe the complex puzzle surrounding the extensive variation in nuclear genome size among eukaryotic species...
- GenomeGenomeIn modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
- Human genomeHuman genomeThe human genome is the genome of Homo sapiens, which is stored on 23 chromosome pairs plus the small mitochondrial DNA. 22 of the 23 chromosomes are autosomal chromosome pairs, while the remaining pair is sex-determining...
- Junk DNA
- Noncoding DNANoncoding DNAIn genetics, noncoding DNA describes components of an organism's DNA sequences that do not encode for protein sequences. In many eukaryotes, a large percentage of an organism's total genome size is noncoding DNA, although the amount of noncoding DNA, and the proportion of coding versus noncoding...
- Plant DNA C-values DatabasePlant DNA C-values DatabaseThe Plant DNA C-values Database is a comprehensive catalogue of C-value data for land plants and algae. The database was created by Prof. Michael D. Bennett and Dr. Ilia J. Leitch of the Royal Botanic Gardens, Kew, UK...
- Selfish DNASelfish DNASelfish DNA refers to those sequences of DNA which, in their purest form, have two distinct properties: the DNA sequence spreads by forming additional copies of itself within the genome; and it makes no specific contribution to the reproductive success of its host organism.This idea was sketched...
- Transposable elements

