
Micro RNA
Encyclopedia

Molecule
A molecule is an electrically neutral group of at least two atoms held together by covalent chemical bonds. Molecules are distinguished from ions by their electrical charge...
found in eukaryotic cell
Eukaryotic Cell
Eukaryotic Cell is an academic journal published by the American Society for Microbiology. The title is commonly abbreviated EC and the ISSN is 1535-9778 for the print version, and 1535-9786 for the electronic version....
s. A microRNA molecule has very few nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
s (an average of 22) compared with other RNAs.
miRNAs are post-transcriptional regulators that bind to complementary sequences on target messenger RNA transcripts
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
(mRNAs), usually resulting in translational repression or target degradation and gene silencing
Gene silencing
Gene silencing is a general term describing epigenetic processes of gene regulation. The term gene silencing is generally used to describe the "switching off" of a gene by a mechanism other than genetic modification...
. The human genome
Genome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
may encode over 1000 miRNAs, which may target about 60% of mammalian genes and are abundant in many human cell types.
miRNAs show very different characteristics between plants and metazoans
Animal
Animals are a major group of multicellular, eukaryotic organisms of the kingdom Animalia or Metazoa. Their body plan eventually becomes fixed as they develop, although some undergo a process of metamorphosis later on in their life. Most animals are motile, meaning they can move spontaneously and...
. In plants the miRNA complementarity to its mRNA target is nearly perfect, with no or few mismatched bases
Base pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
. In metazoans, on the other hand, miRNA complementarity typically encompasses the 5' bases 2-7 of the microRNA, the microRNA seed region, and one miRNA can target many different sites on the same mRNA or on many different mRNAs. Another difference is the location of target sites on mRNAs. In metazoans, the miRNA target sites are in the three prime untranslated region
Three prime untranslated region
In molecular genetics, the three prime untranslated region is a particular section of messenger RNA . It is preceeded by the coding region....
s (3'UTR) of the mRNA. In plants, targets can be located in the 3' UTR but are more often in the coding region
Coding region
The coding region of a gene, also known as the coding sequence or CDS, is that portion of a gene's DNA or RNA, composed of exons, that codes for protein. The region is bounded nearer the 5' end by a start codon and nearer the 3' end with a stop codon...
itself. MiRNAs are well conserved
Conserved sequence
In biology, conserved sequences are similar or identical sequences that occur within nucleic acid sequences , protein sequences, protein structures or polymeric carbohydrates across species or within different molecules produced by the same organism...
in eukaryotic organisms and are thought to be a vital and evolutionarily ancient component of genetic regulation.
The first miRNAs were characterized in the early 1990s. However, miRNAs were not recognized as a distinct class of biological regulators with conserved functions until the early 2000s. Since then, miRNA research has revealed multiple roles in negative regulation (transcript degradation and sequestering, translational suppression) and possible involvement in positive regulation (transcriptional and translational activation). By affecting gene regulation, miRNAs are likely to be involved in most biological processes. Different sets of expressed miRNAs are found in different cell types and tissues
Tissue (biology)
Tissue is a cellular organizational level intermediate between cells and a complete organism. A tissue is an ensemble of cells, not necessarily identical, but from the same origin, that together carry out a specific function. These are called tissues because of their identical functioning...
.
Aberrant expression of miRNAs has been implicated in numerous disease states, and miRNA-based therapies are under investigation.
History
MicroRNAs were discovered in 1993 by Victor AmbrosVictor Ambros
Victor Ambros is an American developmental biologist who discovered the first known microRNA . He is a professor at the University of Massachusetts Medical School in Worcester, Massachusetts.-Background:...
, Rosalind Lee and Rhonda Feinbaum during a study of the gene lin-14 in C. elegans
Caenorhabditis elegans
Caenorhabditis elegans is a free-living, transparent nematode , about 1 mm in length, which lives in temperate soil environments. Research into the molecular and developmental biology of C. elegans was begun in 1974 by Sydney Brenner and it has since been used extensively as a model...
development. They found that LIN-14 protein abundance was regulated by a short RNA product encoded by the lin-4 gene. A 61-nucleotide precursor from the lin-4 gene matured to a 22-nucleotide RNA that contained sequences partially complementary to multiple sequences in the 3’ UTR of the lin-14 mRNA. This complementarity was both necessary and sufficient to inhibit the translation of the lin-14 mRNA into the LIN-14 protein. Retrospectively, the lin-4 small RNA was the first microRNA to be identified, though at the time, it was thought to be a nematode idiosyncrasy. Only in 2000 was a second RNA characterized: let-7, which repressed lin-41, lin-14, lin-28, lin-42, and daf-12 expression during developmental stage transitions in C. elegans. let-7 was soon found to be conserved in many species, indicating the existence of a wider phenomenon.
Nomenclature
Under a standard nomenclature system, names are assigned to experimentally confirmed miRNAs before publication of their discovery. The prefix "mir" is followed by a dash and a number, the latter often indicating order of naming. For example, mir-123 was named and likely discovered prior to mir-456. The uncapitalized "mir-" refers to the pre-miRNA, while a capitalized "miR-" refers to the mature form. miRNAs with nearly identical sequences bar one or two nucleotides are annotated with an additional lower case letter. For example, miR-123a would be closely related to miR-123b. Pre-miRNAs that lead to 100% identical mature miRNAs but that are located at different places in the genome are indicated with an additional dash-number suffix. For example, the pre-miRNAs hsa-mir-194-1 and hsa-mir-194-2 lead to an identical mature miRNA (hsa-miR-194) but are located in different regions of the genome. Species of origin is designated with a three-letter prefix, e.g., hsa-miR-123 is a human (Homo sapiens)miRNA and oar-miR-123 is a sheep (Ovis aries) miRNA. Other common prefixes include 'v' for viral (miRNA encoded by a viral genome) and 'd' for Drosophila miRNA (a fruit fly commonly studied in genetic research). When two mature microRNAs originate from opposite arms of the same pre-miRNA, they are denoted with a -3p or -5p suffix. (In the past, this distinction was also made with 's' (sense) and 'as' (antisense)). When relative expression levels are known, an asterisk following the name indicates an miRNA expressed at low levels relative to the miRNA in the opposite arm of a hairpin. For example, miR-123 and miR-123* would share a pre-miRNA hairpin, but more miR-123 would be found in the cell.Biogenesis
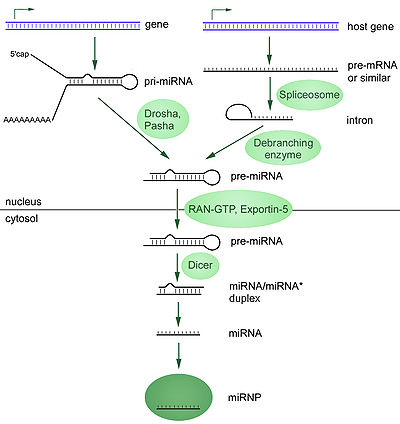
The majority of the characterized miRNA genes are intergenic or oriented antisense to neighboring genes and are therefore suspected to be transcribed as independent units As much as 40% of miRNA genes may lie in the intron
Intron
An intron is any nucleotide sequence within a gene that is removed by RNA splicing to generate the final mature RNA product of a gene. The term intron refers to both the DNA sequence within a gene, and the corresponding sequence in RNA transcripts. Sequences that are joined together in the final...
s of protein and non-protein coding genes or even in exon
Exon
An exon is a nucleic acid sequence that is represented in the mature form of an RNA molecule either after portions of a precursor RNA have been removed by cis-splicing or when two or more precursor RNA molecules have been ligated by trans-splicing. The mature RNA molecule can be a messenger RNA...
s of long nonprotein-coding transcripts. These are usually, though not exclusively, found in a sense orientation. and thus usually are regulated together with their host genes. Other miRNA genes showing a common promoter include the 42-48% of all miRNAs originating from polycistronic units containing multiple discrete loops from which mature miRNAs are processed, although this does not necessarily mean the mature miRNAs of a family will be homologous in structure and function. The promoters mentioned have been shown to have some similarities in their motifs to promoters of other genes transcribed by RNA polymerase II such as protein coding genes. The DNA template is not the final word on mature miRNA production: 6% of human miRNAs show RNA editing ( IsomiRs), the site-specific modification of RNA sequences to yield products different from those encoded by their DNA. This increases the diversity and scope of miRNA action beyond that implicated from the genome alone.
Transcription
miRNA genes are usually transcribed by RNA polymerase IIRNA polymerase II
RNA polymerase II is an enzyme found in eukaryotic cells. It catalyzes the transcription of DNA to synthesize precursors of mRNA and most snRNA and microRNA. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase...
(Pol II). The polymerase often binds to a promoter found near the DNA sequence encoding what will become the hairpin loop of the pre-miRNA. The resulting transcript is capped
5' cap
The 5' cap is a specially altered nucleotide on the 5' end of precursor messenger RNA and some other primary RNA transcripts as found in eukaryotes. The process of 5' capping is vital to creating mature messenger RNA, which is then able to undergo translation...
with a specially-modified nucleotide at the 5’ end, polyadenylated
Polyadenylation
Polyadenylation is the addition of a poly tail to an RNA molecule. The poly tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eukaryotes, polyadenylation is part of the process that produces mature messenger RNA for translation...
with multiple adenosines
Adenosine monophosphate
Adenosine monophosphate , also known as 5'-adenylic acid, is a nucleotide that is used as a monomer in RNA. It is an ester of phosphoric acid and the nucleoside adenosine. AMP consists of a phosphate group, the sugar ribose, and the nucleobase adenine...
(a poly(A) tail), and spliced
RNA splicing
In molecular biology and genetics, splicing is a modification of an RNA after transcription, in which introns are removed and exons are joined. This is needed for the typical eukaryotic messenger RNA before it can be used to produce a correct protein through translation...
. Animal miRNAs are initially transcribed as part of one arm of an ∼80 nucleotide RNA stem-loop
Stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions,...
that in turn forms part of a several hundred nucleotides long miRNA precursor termed a primary miRNA (pri-miRNA)s. When a stem-loop precursor is found in the 3’ UTR, a transcript may serve as a pri-miRNA and a mRNA. RNA polymerase III
RNA polymerase III
RNA polymerase III transcribes DNA to synthesize ribosomal 5S rRNA, tRNA and other small RNAs. The genes transcribed by RNA Pol III fall in the category of "housekeeping" genes whose expression is required in all cell types and most environmental conditions...
(Pol III) transcribes some miRNAs, especially those with upstream Alu sequence
Alu sequence
An Alu element is a short stretch of DNA originally characterized by the action of the Alu restriction endonuclease. Alu elements of different kinds occur in large numbers in primate genomes. In fact, Alu elements are the most abundant mobile elements in the human genome. They are derived from the...
s, transfer RNA
Transfer RNA
Transfer RNA is an adaptor molecule composed of RNA, typically 73 to 93 nucleotides in length, that is used in biology to bridge the three-letter genetic code in messenger RNA with the twenty-letter code of amino acids in proteins. The role of tRNA as an adaptor is best understood by...
s (tRNAs), and mammalian wide interspersed repeat (MWIR) promoter units.
Nuclear processing
A single pri-miRNA may contain from one to six miRNA precursors. These hairpin loop structures are composed of about 70 nucleotides each. Each hairpin is flanked by sequences necessary for efficient processing. The double-stranded RNA structure of the hairpins in a pri-miRNA is recognized by a nuclear protein known as DiGeorge Syndrome Critical Region 8Pasha (protein)
Pasha , also known as DGCR8 in vertebrates organisms, is a protein localized to the cell nucleus that is required for microRNA processing. It binds to Drosha, an RNase III enzyme, to form the Microprocessor complex that cleaves a primary transcript known as pri-miRNA to a characteristic stem-loop...
(DGCR8 or "Pasha" in invertebrates), named for its association with DiGeorge Syndrome
DiGeorge syndrome
22q11.2 deletion syndrome, which has several presentations including DiGeorge syndrome , DiGeorge anomaly, velo-cardio-facial syndrome, Shprintzen syndrome, conotruncal anomaly face syndrome, Strong syndrome, congenital thymic aplasia, and thymic hypoplasia is a syndrome caused by the deletion of a...
. DGCR8 associates with the enzyme Drosha
Drosha
Drosha is a Class 2 RNase III enzyme responsible for initiating the processing of microRNA , or short RNA molecules naturally expressed by the cell that regulate a wide variety of other genes by interacting with the RNA-induced silencing complex to induce cleavage of complementary messenger RNA ...
, a protein that cuts RNA, to form the "Microprocessor" complex. In this complex, DGCR8 orients the catalytic RNase III domain of Drosha to liberate hairpins from pri-miRNAs by cleaving RNA about eleven nucleotides from the hairpin base (two helical RNA turns into the stem). The product resulting has a two-nucleotide overhang at its 3’ end; it has 3' hydroxyl and 5' phosphate groups. It is often termed as a pre-miRNA (precursor-miRNA).
pre-miRNAs that are spliced
RNA splicing
In molecular biology and genetics, splicing is a modification of an RNA after transcription, in which introns are removed and exons are joined. This is needed for the typical eukaryotic messenger RNA before it can be used to produce a correct protein through translation...
directly out of intron
Intron
An intron is any nucleotide sequence within a gene that is removed by RNA splicing to generate the final mature RNA product of a gene. The term intron refers to both the DNA sequence within a gene, and the corresponding sequence in RNA transcripts. Sequences that are joined together in the final...
s, bypassing the Microprocessor complex, are known as "Mirtrons
Mirtrons
Mirtrons are a type of microRNAs that are located in the introns of the mRNA encoding host genes. All the miRNAs in plants are derived from the sequential DCL1 cleavages from pri-miRNA to give pre-miRNA , but the mirtrons bypass the DCL1 cleavage and enter as pre-miRNA in the miRNA maturation...
." Originally thought to exist only in Drosophila and C. elegans, mirtrons have now been found in mammals.
Perhaps as many as 16% of pri-miRNAs may be altered through nuclear RNA editing
RNA editing
The term RNA editing describes those molecular processes in which the information content in an RNA molecule is altered through a chemical change in the base makeup. To date, such changes have been observed in tRNA, rRNA, mRNA and microRNA molecules of eukaryotes but not prokaryotes...
. Most commonly, enzymes known as adenosine deaminases acting on RNA (ADARs) catalyze adenosine to inosine
Inosine
Inosine is a nucleoside that is formed when hypoxanthine is attached to a ribose ring via a β-N9-glycosidic bond....
(A to I) transitions. RNA editing can halt nuclear processing (for example, of pri-miR-142, leading to degradation by the ribonuclease Tudor-SN) and alter downstream processes including cytoplasmic miRNA processing and target specificity (e.g., by changing the seed region of miR-376 in the central nervous system).
Nuclear export
pre-miRNA hairpins are exported from the nucleus in a process involving the nucleocytoplasmic shuttle Exportin-5XPO5
Exportin-5 is a protein that in humans is encoded by the XPO5 gene.-Interactions:XPO5 has been shown to interact with ILF3 and Ran .-Further reading:...
. This protein, a member of the karyopherin
Karyopherin
Karyopherins are a group of proteins involved in transporting molecules from the cytoplasm into the nucleus of eukaryotic cells. The inside of the nucleus is called the karyoplasm . Generally, karyopherin-mediated transport occurs through the nuclear pore, which acts as a gateway into and out of...
family, recognizes a two-nucleotide overhang left by the RNase III enzyme Drosha at the 3' end of the pre-miRNA hairpin. Exportin-5-mediated transport to the cytoplasm is energy-dependent, using GTP bound to the Ran
Ran (biology)
Ran is a small 25Kda protein that is involved in transport into and out of the cell nucleus during interphase and also involved in mitosis. It is a member of the Ras superfamily....
protein.
Cytoplasmic processing
In cytoplasmCytoplasm
The cytoplasm is a small gel-like substance residing between the cell membrane holding all the cell's internal sub-structures , except for the nucleus. All the contents of the cells of prokaryote organisms are contained within the cytoplasm...
, the pre-miRNA hairpin is cleaved by the RNase III enzyme Dicer. This endoribonuclease interacts with the 3' end of the hairpin and cuts away the loop joining the 3' and 5' arms, yielding an imperfect miRNA:miRNA* duplex about 22 nucleotides in length. Overall hairpin length and loop size influence the efficiency of Dicer processing, and the imperfect nature of the miRNA:miRNA* pairing also affects cleavage. Although either strand of the duplex may potentially act as a functional miRNA, only one strand is usually incorporated into the RNA-induced silencing complex (RISC) where the miRNA and its mRNA target interact.
Biogenesis in plants
miRNA biogenesis in plants differs from metazoan biogenesis mainly in the steps of nuclear processing and export. Instead of being cleaved by two different enzymes, once inside and once outside the nucleus, both cleavages of the plant miRNA is performed by a Dicer homolog, called Dicer-like1 (DL1). DL1 is only expressed in the nucleus of plant cells, which indicates that both reactions take place inside the nucleus. Before plant miRNA:miRNA* duplexes are transported out of the nucleus its 3' overhangs are methylated by a RNA methyltransferaseMethyltransferase
A methyltransferase is a type of transferase enzyme that transfers a methyl group from a donor to an acceptor.Methylation often occurs on nucleic bases in DNA or amino acids in protein structures...
protein called Hua-Enhancer1
Hua-Enhancer1
Hua-Enhancer1 is a RNA methyltransferase protein that plays a role in the maturation of miRNA in plants. HE1 methylates the 3' overhangs of miRNA:miRNA* duplexes before they are exported out of the nucleus to the cytoplasm....
(HEN1). The duplex is then transported out of the nucleus to the cytoplasm by a protein called Hasty (HST), an Exportin 5 homolog, where they disassemble and the mature miRNA is incorporated into the RISC.
The RNA-induced silencing complex
The mature miRNA is part of an active RNA-induced silencing complex (RISC) containing Dicer and many associated proteins. RISC is also known as a microRNA ribonucleoprotein complex (miRNP); RISC with incorporated miRNA is sometimes referred to as "miRISC."Dicer processing of the pre-miRNA is thought to be coupled with unwinding of the duplex. Generally, only one strand is incorporated into the miRISC, selected on the basis of its thermodynamic instability and weaker base-pairing relative to the other strand. The position of the stem-loop may also influence strand choice. The other strand, called the passenger strand due to its lower levels in the steady state, is denoted with an asterisk (*) and is normally degraded. In some cases, both strands of the duplex are viable and become functional miRNA that target different mRNA populations.
Members of the argonaute (Ago) protein family are central to RISC function. Argonautes are needed for miRNA-induced silencing and contain two conserved RNA binding domains: a PAZ domain that can bind the single stranded 3’ end of the mature miRNA and a PIWI domain that structurally resembles ribonuclease-H and functions to interact with the 5’ end of the guide strand. They bind the mature miRNA and orient it for interaction with a target mRNA. Some argonautes, for example human Ago2, cleave target transcripts directly; argonautes may also recruit additional proteins to achieve translational repression. The human genome encodes eight argonaute proteins divided by sequence similarities into two families: AGO (with four members present in all mammalian cells and called E1F2C/hAgo in humans), and PIWI (found in the germ line and hematopoietic stem cells).
Additional RISC components include TRBP
TARBP2
RISC-loading complex subunit TARBP2 is a protein that in humans is encoded by the TARBP2 gene.-Interactions:TARBP2 has been shown to interact with Protein kinase R and RBM14.-Further reading:...
[human immunodeficiency virus (HIV) transactivating response RNA (TAR) binding protein], PACT (protein activator of the interferon induced protein kinase (PACT), the SMN complex, fragile X mental retardation protein (FMRP), and Tudor staphylococcal nuclease-domain-containing protein (Tudor-SN).
Mode of Silencing
Gene silencing may occur either via mRNA degradation or preventing mRNA from being translated. It has been demonstrated that if there is complete complementation between the miRNA and target mRNA sequence, Ago2 can cleave the mRNA and lead to direct mRNA degradation. Yet, if there isn't complete complementation the silencing is achieved by preventing translation.miRNA turnover
Turnover of mature miRNA is needed for rapid changes in miRNA expression profiles. During miRNA maturation in the cytoplasm, uptake by the Argonaute protein is thought to stabilize the guide strand, while the opposite (* or "passenger") strand is preferentially destroyed. In what has been called a "Use it or lose it" strategy, Argonaute may preferentially retain miRNAs with many targets over miRNAs with few or no targets, leading to degradation of the non-targeting molecules.Decay of mature miRNAs in Caenorhabditis elegans
Caenorhabditis elegans
Caenorhabditis elegans is a free-living, transparent nematode , about 1 mm in length, which lives in temperate soil environments. Research into the molecular and developmental biology of C. elegans was begun in 1974 by Sydney Brenner and it has since been used extensively as a model...
is mediated by the 5´-to-3´ exoribonuclease XRN2
XRN2
5'-3' Exoribonuclease 2 also known as DHM1-like protein is an exoribonuclease enzyme that in humans is encoded by the XRN2 gene....
, also known as Rat1p. In plants, SDN (small RNA degrading nuclease) family members degrade miRNAs in the opposite (3'-to-5') direction. Similar enzymes are encoded in animal genomes, but their roles have not yet been described.
Several miRNA modifications affect miRNA stability. As indicated by work in the model organism Arabidopsis thaliana
Arabidopsis thaliana
Arabidopsis thaliana is a small flowering plant native to Europe, Asia, and northwestern Africa. A spring annual with a relatively short life cycle, arabidopsis is popular as a model organism in plant biology and genetics...
(thale cress), mature plant miRNAs appear to be stabilized by the addition of methyl moieties at the 3' end. The 2'-O-conjugated methyl groups block the addition of uracil (U) residues by uridyltransferase enzymes, a modification that may be associated with miRNA degradation. However, uridylation may also protect some miRNAs; the consequences of this modification are incompletely understood. Uridylation of some animal miRNAs has also been reported. Both plant and animal miRNAs may be altered by addition of adenine (A) residues to the 3' end of the miRNA. An extra A added to the end of mammalian miR-122
MiR-122
miR-122 is a miRNA that is conserved between vertebrate species. miR-122 is not present in invertebrates, and no close paralogs of miR-122 have been detected. miR-122 expression is specific to the liver, where it has been implicated as a regulator of fatty-acid metabolism in mouse studies. Reduced...
, a liver-enriched miRNA important in Hepatitis C, stabilizes the molecule, and plant miRNAs ending with an adenine residue have slower decay rates.
Cellular functions
The function of miRNAs appears to be in gene regulation. For that purpose, a miRNA is complementaryComplementarity (molecular biology)
In molecular biology, complementarity is a property of double-stranded nucleic acids such as DNA, as well as DNA:RNA duplexes. Each strand is complementary to the other in that the base pairs between them are non-covalently connected via two or three hydrogen bonds...
to a part of one or more messenger RNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
s (mRNAs). Animal miRNAs are usually complementary to a site in the 3' UTR whereas plant miRNAs are usually complementary to coding regions of mRNAs. Perfect or near perfect base pairing with the target RNA promotes cleavage of the RNA. This is the primary mode of plant miRNAs. In animals miRNAs more often have only partly the right sequence of nucleotides to bond with the target mRNA. The match-ups are imperfect. Animal miRNAs inhibit protein translation of the target mRNA (this exists in plants as well but is less common). MicroRNAs that are partially complementary to a target can also speed up deadenylation, causing mRNAs to be degraded sooner. For partially complementary microRNAs to recognise their targets, nucleotides 2–7 of the miRNA (its 'seed region') still have to be perfectly complementary. miRNAs occasionally also cause histone modification and DNA methylation
DNA methylation
DNA methylation is a biochemical process that is important for normal development in higher organisms. It involves the addition of a methyl group to the 5 position of the cytosine pyrimidine ring or the number 6 nitrogen of the adenine purine ring...
of promoter sites, which affects the expression of target genes.
Unlike plant microRNAs, the animal microRNAs target a diverse set of genes. However, genes involved in functions common to all cells, such as gene expression, have relatively fewer microRNA target sites and seem to be under selection to avoid targeting by microRNAs.
dsRNA can also activate gene expression
Gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA , transfer RNA or small nuclear RNA genes, the product is a functional RNA...
, a mechanism that has been termed "small RNA-induced gene activation" or RNAa
RNAa
Small double-stranded RNA has been found to silence gene expression by an evolutionally conserved mechanism known as RNA interference or RNAi. Such dsRNAs are called small interfering RNAs or siRNA. RNAi can occur at both transcriptional and post-transcriptional levels...
. dsRNAs targeting gene promoters can induce potent transcriptional activation of associated genes. This was demonstrated in human cells using synthetic dsRNAs termed small activating RNAs (saRNA
SaRNA
Small activating RNA molecules can induce target gene expression in a phenomenon referred to as dsRNA-induced transcriptional activation ....
s), but has also been demonstrated for endogenous microRNA.
Interactions between microRNAs and complementary sequences on genes and even pseudogene
Pseudogene
Pseudogenes are dysfunctional relatives of known genes that have lost their protein-coding ability or are otherwise no longer expressed in the cell...
s that share sequence homology are thought to be a back channel of communication regulating expression levels between paralogous genes. Given the name "competing endogenous RNAs" (ceRNA
Cerna
Cerna may refer to:Croatia:* Cerna, Vukovar-Syrmia County, a village in Vukovar-Syrmia County, CroatiaRomania:* Cerna River , a river in Romania, tributary of the Danube* Cerna River , a river in Romania, tributary of the Mureş River...
s), these microRNAs bind to "microRNA response elements" on genes and pseudogenes and may provide another explanation for the persistence of non-coding ("junk") DNA.
Evolution
MicroRNAs are significant phylogenetic markers because of their astonishingly low rate of evolution. Their origin may have permitted the development of morphological innovation, and by making gene expression more specific and 'fine-tunable', permitted the genesis of complex organs and perhaps, ultimately, complex life. Indeed, rapid bursts of morphological innovation are generally associated with a high rate of microRNA accumulation.MicroRNAs originate predominantly by the random formation of hairpins in "non-coding" sections of DNA (i.e. introns or intergene regions), but also by the duplication and modification of existing microRNAs. The rate of evolution (i.e. nucleotide substitution) in recently-originated microRNAs is comparable to that elsewhere in the non-coding DNA, implying evolution by neutral drift; however, older microRNAs have a much lower rate of change (often less than one substitution per hundred million years), suggesting that once a microRNA gains a function it undergoes extreme purifying selection. At this point, a microRNA is rarely lost from an animal's genome, although microRNAs which are more recently derived (and thus presumably non-functional) are frequently lost. This makes them a valuable phylogenetic marker, and they are being looked upon as a possible solution to such outstanding phylogenetic problems as the relationships of arthropods.
MicroRNAs feature in the genome
Genome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
s of most eukaryotic organisms, from the brown algae to the metazoa. Across all species, in excess of 5000 had been identified by March 2010. Whilst short RNA sequences (50 – hundreds of base pairs) of a broadly comparable function occur in bacteria, bacteria lack true microRNAs.
Experimental detection and manipulation of miRNA
MicroRNA expression can be quantified in a two-step polymerase chain reaction process of modified RT-PCR followed by quantitative real-time PCR. Variations of this method achieve absolute or relative quantification. miRNAs can also be hybridized to microarrayMicroarray
A microarray is a multiplex lab-on-a-chip. It is a 2D array on a solid substrate that assays large amounts of biological material using high-throughput screening methods.Types of microarrays include:...
s, slides or chips with probes to hundreds or thousands of miRNA targets, so that relative levels of miRNAs can be determined in different samples. MicroRNAs can be both discovered and profiled by high-throughput sequencing methods. The activity of an miRNA can be experimentally inhibited using a locked nucleic acid
Locked nucleic acid
A locked nucleic acid , often referred to as inaccessible RNA, is a modified RNA nucleotide. The ribose moiety of an LNA nucleotide is modified with an extra bridge connecting the 2' oxygen and 4' carbon. The bridge "locks" the ribose in the 3'-endo conformation, which is often found in the A-form...
(LNA) oligo, a Morpholino
Morpholino
In molecular biology, a Morpholino is a molecule in a particular structural family that is used to modify gene expression. Morpholino oligomers are an antisense technology used to block access of other molecules to specific sequences within nucleic acid...
oligo or a 2'-O-methyl RNA oligo. Additionally, a specific miRNA can be silenced by a complementary antagomir
Antagomir
Antagomirs are one of a novel class of chemically engineered oligonucleotides. Antagomirs are used to silence endogenous microRNA.An antagomir is a small synthetic RNA that is perfectly complementary to the specific miRNA target with either mispairing at the cleavage site of Ago2 or some sort of...
. MicroRNA maturation can be inhibited at several points by steric-blocking oligos. The miRNA target site of an mRNA transcript can also be blocked by a steric-blocking oligo. For the “in situ” detection of miRNA, the use of LNA is currently the only efficient method. The locked conformation of LNA results in enhanced hybridization properties and increases sensitivity and selectivity, making it ideal for detection of short miRNA.
miRNA and disease
Just as miRNA is involved in the normal functioning of eukaryotic cells, so has dysregulation of miRNA been associated with disease. A manually curated, publicly available database miR2Disease documents known relationships between miRNA dysregulation and human disease.miRNA and inherited diseases
A mutation in the seed region of miR-96 causes hereditary progressive hearing loss. A mutation in the seed region of miR-184 causes hereditary keratoconus with anterior polar cataract. Deletion of the miR-17~92 cluster causes skeletal and growth defects.miRNA and cancer
Several miRNAs have been found to have links with some types of cancerCancer
Cancer , known medically as a malignant neoplasm, is a large group of different diseases, all involving unregulated cell growth. In cancer, cells divide and grow uncontrollably, forming malignant tumors, and invade nearby parts of the body. The cancer may also spread to more distant parts of the...
. MicroRNA-21
MIRN21
microRNA 21 also known as hsa-mir-21 or miRNA21 is a mammalian microRNA that is encoded by the MIR21 gene.MIRN21 was one of the first mammalian microRNAs identified. The mature miR-21 sequence is strongly conserved throughout evolution...
is one of the first microRNAs that was identified as an oncomiR
Oncomir
An oncomir is a miRNA that is associated to cancer, for example, because it is found to be present in primary tumors or in tumor cell lines....
.
A study of mice altered to produce excess c-Myc
Myc
Myc is a regulator gene that codes for a transcription factor. In the human genome, Myc is located on chromosome 8 and is believed to regulate expression of 15% of all genes through binding on Enhancer Box sequences and recruiting histone acetyltransferases...
— a protein with mutated forms implicated in several cancers — shows that miRNA has an effect on the development of cancer. Mice that were engineered to produce a surplus of types of miRNA found in lymphoma
Lymphoma
Lymphoma is a cancer in the lymphatic cells of the immune system. Typically, lymphomas present as a solid tumor of lymphoid cells. Treatment might involve chemotherapy and in some cases radiotherapy and/or bone marrow transplantation, and can be curable depending on the histology, type, and stage...
cells developed the disease within 50 days and died two weeks later. In contrast, mice without the surplus miRNA lived over 100 days.
Leukemia can be caused by the insertion of a viral genome next to the 17-92 array of microRNAs leading to increased expression of this microRNA.
Another study found that two types of miRNA inhibit the E2F1 protein, which regulates cell proliferation
Cell growth
The term cell growth is used in the contexts of cell development and cell division . When used in the context of cell division, it refers to growth of cell populations, where one cell grows and divides to produce two "daughter cells"...
. miRNA appears to bind to messenger RNA before it can be translated to proteins that switch gene
Gene
A gene is a molecular unit of heredity of a living organism. It is a name given to some stretches of DNA and RNA that code for a type of protein or for an RNA chain that has a function in the organism. Living beings depend on genes, as they specify all proteins and functional RNA chains...
s on and off.
By measuring activity among 217 genes encoding miRNA, patterns of gene activity that can distinguish types of cancers can be discerned. miRNA signatures may enable classification of cancer. This will allow doctors to determine the original tissue type which spawned a cancer and to be able to target a treatment course based on the original tissue type. miRNA profiling has already been able to determine whether patients with chronic lymphocytic leukemia
Chronic lymphocytic leukemia
B-cell chronic lymphocytic leukemia , also known as chronic lymphoid leukemia , is the most common type of leukemia. Leukemias are cancers of the white blood cells . CLL affects B cell lymphocytes. B cells originate in the bone marrow, develop in the lymph nodes, and normally fight infection by...
had slow growing or aggressive forms of the cancer.
Transgenic mice that over-express or lack specific miRNAs have provided insight into the role of small RNAs in various malignancies.
A novel miRNA-profiling based screening assay for the detection of early-stage colorectal cancer
Colorectal cancer
Colorectal cancer, commonly known as bowel cancer, is a cancer caused by uncontrolled cell growth , in the colon, rectum, or vermiform appendix. Colorectal cancer is clinically distinct from anal cancer, which affects the anus....
has been developed and is currently in clinical trials. Early results showed that blood plasma samples collected from patients with early, resectable (Stage II) colorectal cancer could be distinguished from those of sex-and age-matched healthy volunteers. Sufficient selectivity and specificity could be achieved using small (less than 1 mL) samples of blood. The test has potential to be a cost-effective, non-invasive way to identify at-risk patients who should undergo colonoscopy.
Another role for miRNA in cancers is to use their expression level as a prognostic, for example one study on NSCLC samples found that low miR-324a levels could serve as a prognostic indicator of poor survival, another found that either high miR-185 or low miR-133b levels correlated with metastasis
Metastasis
Metastasis, or metastatic disease , is the spread of a disease from one organ or part to another non-adjacent organ or part. It was previously thought that only malignant tumor cells and infections have the capacity to metastasize; however, this is being reconsidered due to new research...
and poor survival in colorectal cancer
Colorectal cancer
Colorectal cancer, commonly known as bowel cancer, is a cancer caused by uncontrolled cell growth , in the colon, rectum, or vermiform appendix. Colorectal cancer is clinically distinct from anal cancer, which affects the anus....
.
miRNA and heart disease
The global role of miRNA function in the heart has been addressed by conditionally inhibiting miRNA maturation in the murine heart, and has revealed that miRNAs play an essential role during its development. miRNA expression profiling studies demonstrate that expression levels of specific miRNAs change in diseased human hearts, pointing to their involvement in cardiomyopathiesCardiomyopathy
Cardiomyopathy, which literally means "heart muscle disease," is the deterioration of the function of the myocardium for any reason. People with cardiomyopathy are often at risk of arrhythmia or sudden cardiac death or both. Cardiomyopathy can often go undetected, making it especially dangerous to...
. Furthermore, studies on specific miRNAs in animal models have identified distinct roles for miRNAs both during heart development and under pathological conditions, including the regulation of key factors important for cardiogenesis, the hypertrophic growth response, and cardiac conductance.
miRNA and the nervous system
miRNAs appear to regulate the nervous systemNervous system
The nervous system is an organ system containing a network of specialized cells called neurons that coordinate the actions of an animal and transmit signals between different parts of its body. In most animals the nervous system consists of two parts, central and peripheral. The central nervous...
. Neural miRNAs are involved at various stages of synaptic development, including dendritogenesis (involving miR-132, miR-134 and miR-124
Mir-124 microRNA precursor family
The miR-124 microRNA precursor is a small non-coding RNA molecule that has been identified in flies , nematode worms , mouse and human . The mature ~21 nucleotide microRNAs are processed from hairpin precursor sequences by the Dicer enzyme, and in this case originates from the 3' arm. miR-124 has...
), synapse
Synapse
In the nervous system, a synapse is a structure that permits a neuron to pass an electrical or chemical signal to another cell...
formation and synapse maturation (where miR-134 and miR-138 are thought to be involved). Some studies find altered miRNA expression in schizophrenia
Schizophrenia
Schizophrenia is a mental disorder characterized by a disintegration of thought processes and of emotional responsiveness. It most commonly manifests itself as auditory hallucinations, paranoid or bizarre delusions, or disorganized speech and thinking, and it is accompanied by significant social...
.
miRNA and non-coding RNAs
When the human genome project mapped its first chromosome in 1999, it was predicted the genome would contain over 100,000 protein coding genes. However, only around 20,000 were eventually identified (International Human Genome Sequencing Consortium, 2004). Since then, the advent of bioinformatics approaches combined with genome tiling studies examining the transcriptome, systematic sequencing of full length cDNA libraries, and experimental validation (including the creation of miRNA derived antisense oligonucleotides called antagomirAntagomir
Antagomirs are one of a novel class of chemically engineered oligonucleotides. Antagomirs are used to silence endogenous microRNA.An antagomir is a small synthetic RNA that is perfectly complementary to the specific miRNA target with either mispairing at the cleavage site of Ago2 or some sort of...
s) have revealed that many transcripts are non protein-coding RNA, including several snoRNAs and miRNAs.
See also
- Gene expressionGene expressionGene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA , transfer RNA or small nuclear RNA genes, the product is a functional RNA...
- RNAiRNAIRNAI is a non-coding RNA that is an antisense repressor of the replication of some E. coli plasmids, including ColE1. Plasmid replication is usually initiated by RNAII, which acts as a primer by binding to its template DNA. The complementary RNAI binds RNAII prohibiting it from its initiation role...
- siRNASírnaSírna Sáeglach , son of Dian mac Demal, son of Demal mac Rothechtaid, son of Rothechtaid mac Main, was, according to medieval Irish legend and historical tradition, a High King of Ireland...
- List of miRNA target prediction tools
- List of miRNA gene prediction tools
Further reading
- This paper discusses the role of microRNAs in viral oncogenesis:
- This paper discusses the role of microRNAs in Host-virus interactions:
- This paper defines miRNA and proposes guidelines to follow in classifying RNA genes as miRNA:
- This paper discusses the processes that miRNA and siRNAs are involved in, in the context of 2 articles in the same issue of the journal Science:
- This paper describes the discovery of lin-4, the first miRNA to be discovered:
- "This paper showes that the over-expression of a miRNA in mice leads to cancer:"
- This paper discusses the role of microRNAs in skin:
External links
- The miRBase database
- The miRNA Blog
- miR2Disease, a manually curated database documenting known relationships between miRNA dysregulation and human disease.

