
Nucleic acid design
Encyclopedia
Nucleic acid design is the process of generating a set of nucleic acid
base sequences that will associate into a desired conformation. Nucleic acid design is central to the fields of DNA nanotechnology
and DNA computing
. It is necessary because there are many possible sequences of nucleic acid strands that will fold into a given secondary structure
, but many of these sequences will have undesired additional interactions which must be avoided. In addition, there are many tertiary structure
considerations which affect the choice of a secondary structure for a given design.
Nucleic acid design has similar goals to protein design
: in both, the sequence of monomers is rationally designed
to favor the desired folded or associated structure and to disfavor alternate structures. However, nucleic acid design has the advantage of being a much computationally simpler problem, since the simplicity of Watson-Crick base pair
ing rules leads to simple heuristic
methods which yield experimentally robust designs. Computational models for protein folding
require tertiary structure information whereas nucleic acid design can operate largely on the level of secondary structure
. However, nucleic acid structures are less versatile than proteins in their functionality.
Nucleic acid design can be considered the inverse of nucleic acid structure prediction
. In structure prediction, the structure is determined from a known sequence, while in nucleic acid design, a sequence is generated which will form a desired structure.
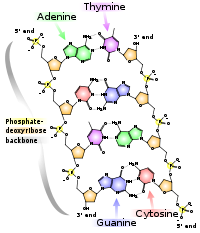 The structure of nucleic acids
The structure of nucleic acids
consists of a sequence of nucleotide
s. There are four types of nucleotides distinguished by which of the four nucleobase
s they contain: in DNA these are adenine
(A), cytosine
(C), guanine
(G), and thymine
(T). Nucleic acids have the property that two molecules will bind to each other to form a double helix only if the two sequences are complementary
, that is, they can form matching sequences of base pair
s. Thus, in nucleic acids the sequence determines the pattern of binding and thus the overall structure.
Nucleic acid design is the process by which, given a desired target structure or functionality, sequences are generated for nucleic acid strands which will self-assemble into that target structure. Nucleic acid design encompasses all levels of nucleic acid structure
:
One of the greatest concerns in nucleic acid design is ensuring that the target structure has the lowest energy (i.e. is the most thermodynamically favorable) whereas misformed structures have higher energy and are thus unfavored.
These goals can be achieved through the use of a number of approaches, including heuristic
, thermodynamic, and geometrical ones. Almost all nucleic acid design tasks are aided by computers, and a number of software packages are available for many of these tasks.
Two considerations in nucleic acid design are that desired hybridizations should have melting temperatures in a narrow range, and any spurious interactions should have very low melting temperatures (i.e. they should be very weak). There is also a contrast between affinity-optimizing "positive design", seeks to minimize the energy of the desired structure in an absolute sense, and specificity-optimizing "negative design", which considers the energy of the target structure relative to those of undesired structures. Algorithms which implement both kinds of design tend to perform better than those that consider only one type.
methods use simple criteria which can be quickly evaluated to judge the suitability of different sequences for a given secondary structure. They have the advantage of being much less computationally expensive than the energy minimization
algorithms needed for thermodynamic or geometrical modeling, and being easier to implement, but at the cost of being less rigorous than these models.
Sequence symmetry minimization is the oldest approach to nucleic acid design and was first used to design immobile versions of branched DNA structures. Sequence symmetry minimization divides the nucleic acid sequence into overlapping subsequences of a fixed length, called the criterion length. Each of the 4N possible subsequences of length N is allowed to appear only once in the sequence. This ensures that no undesired hybridizations can occur which have a length greater than or equal to the criterion length.
A related heuristic approach is to consider the "mismatch distance", meaning the number of positions in a certain frame where the bases are not complementary
. A greater mismatch distance lessens the chance that a strong spurious interaction can happen. This is related to the concept of Hamming distance
in information theory
. Another related but more involved approach is to use methods from coding theory
to construct nucleic acid sequences with desired properties.
of a nucleic acid complex along with its sequence can be used to predict the thermodynamic
properties of the complex.
When thermodynamic models are used in nucleic acid design, there are usually two considerations: desired hybridizations should have melting temperatures in a narrow range, and any spurious interactions should have very low melting temperatures (i.e. they should be very weak). The Gibbs free energy
of a perfectly matched nucleic acid duplex can be predicted using a nearest neighbor model. This model considers only the interactions between a nucleotide and its nearest neighbors on the nucleic acid strand, by summing the free energy of each of the overlapping two-nucleotide subwords of the duplex. This is then corrected for self-complementary monomers and for GC-content
. Once the free energy is known, the melting temperature
of the duplex can be determined. GC-content alone can also be used to estimate the free energy and melting temperature of a nucleic acid duplex. This is less accurate but also much less computationally costly.
Software for thermodynamic modeling of nucleic acids includes Nupack,
mfold/UNAFold, and Vienna.
A related approach, inverse secondary structure prediction, uses stochastic
local search which improves a nucleic acid sequence by running a structure prediction
algorithm and the modifying the sequence to eliminate unwanted features.
. This is important because designed nucleic acid complexes usually contain multiple junction points, which introduces geometric constraints to the system. These constraints stem from the basic structure of nucleic acids
, mainly that the double helix formed by nucleic acid duplexes has a fixed helicity of about 10.4 base pair
s per turn, and is relatively stiff
. Because of these constraints, the nucleic acid complexes are sensitive to the relative orientation of the major and minor grooves at junction points. Geometrical modeling can detect strain
stemming from misalignments in the structure, which can then be corrected by the designer.
Geometric models of nucleic acids for DNA nanotechnology
generally use reduced representations of the nucleic acid, because simulating every atom would be very computationally expensive for such large systems. Models with three pseudo-atoms per base pair, representing the two backbone sugars and the helix axis, have been reported to have a sufficient level of detail to predict experimental results. However, models with five pseudo-atoms per base pair, explicitly including the backbone phosphates, are also used.
Software for geometrical modeling of nucleic acids includes GIDEON,
Tiamat,
Nanoengineer-1,
and UNIQUIMER 3D.
Geometrical concerns are especially of interest in the design of DNA origami
, because the sequence is predetermined by the choice of scaffold strand. Software specifically for DNA origami design has been made, including caDNAno
and SARSE.
to design strands which will self-assemble into a desired target structure. These include examples such as DNA machine
s, periodic two- and three-dimensional lattices, polyhedra, and DNA origami
. It can also be used to create sets of nucleic acid strands which are "orthogonal", or non-interacting with each other, so as to minimize or eliminate spurious interactions. This is useful in DNA computing
, as well as for molecular barcoding applications in chemical biology
and biotechnology
.
—A comparison and evaluation of a number of heuristic and thermodynamic methods for nucleic acid design.
—One of the earliest papers on nucleic acid design, describing the use of sequence symmetry minimization to construct immoble branched junctions.
—A review comparing the capabilities of available nucleic acid design software.
Nucleic acid
Nucleic acids are biological molecules essential for life, and include DNA and RNA . Together with proteins, nucleic acids make up the most important macromolecules; each is found in abundance in all living things, where they function in encoding, transmitting and expressing genetic information...
base sequences that will associate into a desired conformation. Nucleic acid design is central to the fields of DNA nanotechnology
DNA nanotechnology
DNA nanotechnology is a branch of nanotechnology which uses the molecular recognition properties of DNA and other nucleic acids to create designed, artificial structures out of DNA for technological purposes. In this field, DNA is used as a structural material rather than as a carrier of genetic...
and DNA computing
DNA computing
DNA computing is a form of computing which uses DNA, biochemistry and molecular biology, instead of the traditional silicon-based computer technologies. DNA computing, or, more generally, biomolecular computing, is a fast developing interdisciplinary area...
. It is necessary because there are many possible sequences of nucleic acid strands that will fold into a given secondary structure
Nucleic acid secondary structure
The secondary structure of a nucleic acid molecule refers to the basepairing interactions within a single molecule or set of interacting molecules, and can be represented as a list of bases which are paired in a nucleic acid molecule....
, but many of these sequences will have undesired additional interactions which must be avoided. In addition, there are many tertiary structure
Nucleic acid tertiary structure
300px|thumb|upright|alt = Colored dice with checkered background|Example of a large catalytic RNA. The self-splicing group II intron from Oceanobacillus iheyensis....
considerations which affect the choice of a secondary structure for a given design.
Nucleic acid design has similar goals to protein design
Protein design
Protein design is the design of new protein molecules, either from scratch or by making calculated variations on a known structure. The use of rational design techniques for proteins is a major aspect of protein engineering....
: in both, the sequence of monomers is rationally designed
Rational design
In chemical biology and biomolecular engineering, rational design is the strategy of creating new molecules with a certain functionality, based upon the ability to predict how the molecule's structure will affect its behavior through physical models...
to favor the desired folded or associated structure and to disfavor alternate structures. However, nucleic acid design has the advantage of being a much computationally simpler problem, since the simplicity of Watson-Crick base pair
Base pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
ing rules leads to simple heuristic
Heuristic
Heuristic refers to experience-based techniques for problem solving, learning, and discovery. Heuristic methods are used to speed up the process of finding a satisfactory solution, where an exhaustive search is impractical...
methods which yield experimentally robust designs. Computational models for protein folding
Protein folding
Protein folding is the process by which a protein structure assumes its functional shape or conformation. It is the physical process by which a polypeptide folds into its characteristic and functional three-dimensional structure from random coil....
require tertiary structure information whereas nucleic acid design can operate largely on the level of secondary structure
Nucleic acid secondary structure
The secondary structure of a nucleic acid molecule refers to the basepairing interactions within a single molecule or set of interacting molecules, and can be represented as a list of bases which are paired in a nucleic acid molecule....
. However, nucleic acid structures are less versatile than proteins in their functionality.
Nucleic acid design can be considered the inverse of nucleic acid structure prediction
Nucleic acid structure prediction
Nucleic acid structure prediction is a computational method to determine nucleic acid secondary and tertiary structure from its sequence. Secondary structure can be predicted from a single or from several nucleic acid sequences...
. In structure prediction, the structure is determined from a known sequence, while in nucleic acid design, a sequence is generated which will form a desired structure.
Fundamental concepts

Nucleic acid structure
Nucleic acid structure refers to the structure of nucleic acids such as DNA and RNA It is often divided into four different levels:* Primary structure—the raw sequence of nucleobases of each of the component DNA strands;...
consists of a sequence of nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
s. There are four types of nucleotides distinguished by which of the four nucleobase
Nucleobase
Nucleobases are a group of nitrogen-based molecules that are required to form nucleotides, the basic building blocks of DNA and RNA. Nucleobases provide the molecular structure necessary for the hydrogen bonding of complementary DNA and RNA strands, and are key components in the formation of stable...
s they contain: in DNA these are adenine
Adenine
Adenine is a nucleobase with a variety of roles in biochemistry including cellular respiration, in the form of both the energy-rich adenosine triphosphate and the cofactors nicotinamide adenine dinucleotide and flavin adenine dinucleotide , and protein synthesis, as a chemical component of DNA...
(A), cytosine
Cytosine
Cytosine is one of the four main bases found in DNA and RNA, along with adenine, guanine, and thymine . It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached . The nucleoside of cytosine is cytidine...
(C), guanine
Guanine
Guanine is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine . In DNA, guanine is paired with cytosine. With the formula C5H5N5O, guanine is a derivative of purine, consisting of a fused pyrimidine-imidazole ring system with...
(G), and thymine
Thymine
Thymine is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine nucleobase. As the name suggests, thymine may be derived by methylation of uracil at...
(T). Nucleic acids have the property that two molecules will bind to each other to form a double helix only if the two sequences are complementary
Complementarity (molecular biology)
In molecular biology, complementarity is a property of double-stranded nucleic acids such as DNA, as well as DNA:RNA duplexes. Each strand is complementary to the other in that the base pairs between them are non-covalently connected via two or three hydrogen bonds...
, that is, they can form matching sequences of base pair
Base pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
s. Thus, in nucleic acids the sequence determines the pattern of binding and thus the overall structure.
Nucleic acid design is the process by which, given a desired target structure or functionality, sequences are generated for nucleic acid strands which will self-assemble into that target structure. Nucleic acid design encompasses all levels of nucleic acid structure
DNA structure
In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA and RNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, and is a fundamental component in determining its...
:
- Primary structure—the raw sequence of nucleobaseNucleobaseNucleobases are a group of nitrogen-based molecules that are required to form nucleotides, the basic building blocks of DNA and RNA. Nucleobases provide the molecular structure necessary for the hydrogen bonding of complementary DNA and RNA strands, and are key components in the formation of stable...
s of each of the component nucleic acid strands; - Secondary structureNucleic acid secondary structureThe secondary structure of a nucleic acid molecule refers to the basepairing interactions within a single molecule or set of interacting molecules, and can be represented as a list of bases which are paired in a nucleic acid molecule....
—the set of interactions between bases, i.e., which parts of which strands are bound to each other; and - Tertiary structureNucleic acid tertiary structure300px|thumb|upright|alt = Colored dice with checkered background|Example of a large catalytic RNA. The self-splicing group II intron from Oceanobacillus iheyensis....
—the locations of the atoms in three-dimensional space, taking into consideration geometrical and stericSteric effectsSteric effects arise from the fact that each atom within a molecule occupies a certain amount of space. If atoms are brought too close together, there is an associated cost in energy due to overlapping electron clouds , and this may affect the molecule's preferred shape and reactivity.-Steric...
constraints.
One of the greatest concerns in nucleic acid design is ensuring that the target structure has the lowest energy (i.e. is the most thermodynamically favorable) whereas misformed structures have higher energy and are thus unfavored.
These goals can be achieved through the use of a number of approaches, including heuristic
Heuristic
Heuristic refers to experience-based techniques for problem solving, learning, and discovery. Heuristic methods are used to speed up the process of finding a satisfactory solution, where an exhaustive search is impractical...
, thermodynamic, and geometrical ones. Almost all nucleic acid design tasks are aided by computers, and a number of software packages are available for many of these tasks.
Two considerations in nucleic acid design are that desired hybridizations should have melting temperatures in a narrow range, and any spurious interactions should have very low melting temperatures (i.e. they should be very weak). There is also a contrast between affinity-optimizing "positive design", seeks to minimize the energy of the desired structure in an absolute sense, and specificity-optimizing "negative design", which considers the energy of the target structure relative to those of undesired structures. Algorithms which implement both kinds of design tend to perform better than those that consider only one type.
Heuristic methods
HeuristicHeuristic
Heuristic refers to experience-based techniques for problem solving, learning, and discovery. Heuristic methods are used to speed up the process of finding a satisfactory solution, where an exhaustive search is impractical...
methods use simple criteria which can be quickly evaluated to judge the suitability of different sequences for a given secondary structure. They have the advantage of being much less computationally expensive than the energy minimization
Energy minimization
In computational chemistry, energy minimization methods are used to compute the equilibrium configuration of molecules and solids....
algorithms needed for thermodynamic or geometrical modeling, and being easier to implement, but at the cost of being less rigorous than these models.
Sequence symmetry minimization is the oldest approach to nucleic acid design and was first used to design immobile versions of branched DNA structures. Sequence symmetry minimization divides the nucleic acid sequence into overlapping subsequences of a fixed length, called the criterion length. Each of the 4N possible subsequences of length N is allowed to appear only once in the sequence. This ensures that no undesired hybridizations can occur which have a length greater than or equal to the criterion length.
A related heuristic approach is to consider the "mismatch distance", meaning the number of positions in a certain frame where the bases are not complementary
Complementarity (molecular biology)
In molecular biology, complementarity is a property of double-stranded nucleic acids such as DNA, as well as DNA:RNA duplexes. Each strand is complementary to the other in that the base pairs between them are non-covalently connected via two or three hydrogen bonds...
. A greater mismatch distance lessens the chance that a strong spurious interaction can happen. This is related to the concept of Hamming distance
Hamming distance
In information theory, the Hamming distance between two strings of equal length is the number of positions at which the corresponding symbols are different...
in information theory
Information theory
Information theory is a branch of applied mathematics and electrical engineering involving the quantification of information. Information theory was developed by Claude E. Shannon to find fundamental limits on signal processing operations such as compressing data and on reliably storing and...
. Another related but more involved approach is to use methods from coding theory
Coding theory
Coding theory is the study of the properties of codes and their fitness for a specific application. Codes are used for data compression, cryptography, error-correction and more recently also for network coding...
to construct nucleic acid sequences with desired properties.
Thermodynamic models
Information about the secondary structureNucleic acid secondary structure
The secondary structure of a nucleic acid molecule refers to the basepairing interactions within a single molecule or set of interacting molecules, and can be represented as a list of bases which are paired in a nucleic acid molecule....
of a nucleic acid complex along with its sequence can be used to predict the thermodynamic
Thermodynamics
Thermodynamics is a physical science that studies the effects on material bodies, and on radiation in regions of space, of transfer of heat and of work done on or by the bodies or radiation...
properties of the complex.
When thermodynamic models are used in nucleic acid design, there are usually two considerations: desired hybridizations should have melting temperatures in a narrow range, and any spurious interactions should have very low melting temperatures (i.e. they should be very weak). The Gibbs free energy
Gibbs free energy
In thermodynamics, the Gibbs free energy is a thermodynamic potential that measures the "useful" or process-initiating work obtainable from a thermodynamic system at a constant temperature and pressure...
of a perfectly matched nucleic acid duplex can be predicted using a nearest neighbor model. This model considers only the interactions between a nucleotide and its nearest neighbors on the nucleic acid strand, by summing the free energy of each of the overlapping two-nucleotide subwords of the duplex. This is then corrected for self-complementary monomers and for GC-content
GC-content
In molecular biology and genetics, GC-content is the percentage of nitrogenous bases on a DNA molecule that are either guanine or cytosine . This may refer to a specific fragment of DNA or RNA, or that of the whole genome...
. Once the free energy is known, the melting temperature
Melting point
The melting point of a solid is the temperature at which it changes state from solid to liquid. At the melting point the solid and liquid phase exist in equilibrium. The melting point of a substance depends on pressure and is usually specified at standard atmospheric pressure...
of the duplex can be determined. GC-content alone can also be used to estimate the free energy and melting temperature of a nucleic acid duplex. This is less accurate but also much less computationally costly.
Software for thermodynamic modeling of nucleic acids includes Nupack,
mfold/UNAFold, and Vienna.
A related approach, inverse secondary structure prediction, uses stochastic
Stochastic
Stochastic refers to systems whose behaviour is intrinsically non-deterministic. A stochastic process is one whose behavior is non-deterministic, in that a system's subsequent state is determined both by the process's predictable actions and by a random element. However, according to M. Kac and E...
local search which improves a nucleic acid sequence by running a structure prediction
Nucleic acid structure prediction
Nucleic acid structure prediction is a computational method to determine nucleic acid secondary and tertiary structure from its sequence. Secondary structure can be predicted from a single or from several nucleic acid sequences...
algorithm and the modifying the sequence to eliminate unwanted features.
Geometrical models
Geometrical models of nucleic acids are used to predict tertiary structureNucleic acid tertiary structure
300px|thumb|upright|alt = Colored dice with checkered background|Example of a large catalytic RNA. The self-splicing group II intron from Oceanobacillus iheyensis....
. This is important because designed nucleic acid complexes usually contain multiple junction points, which introduces geometric constraints to the system. These constraints stem from the basic structure of nucleic acids
Nucleic acid structure
Nucleic acid structure refers to the structure of nucleic acids such as DNA and RNA It is often divided into four different levels:* Primary structure—the raw sequence of nucleobases of each of the component DNA strands;...
, mainly that the double helix formed by nucleic acid duplexes has a fixed helicity of about 10.4 base pair
Base pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
s per turn, and is relatively stiff
Persistence length
The persistence length is a basic mechanical property quantifying the stiffness of a polymer or of a string.Informally, for pieces of the polymer that are shorter than the persistence length, the molecule behaves rather like a flexible elastic rod, while for pieces of the polymer that are much...
. Because of these constraints, the nucleic acid complexes are sensitive to the relative orientation of the major and minor grooves at junction points. Geometrical modeling can detect strain
Strain (chemistry)
In chemistry, a molecule experiences strain when its chemical structure undergoes some stress which raises its internal energy in comparison to a strain-free reference compound. The internal energy of a molecule consists of all the energy stored within it. A strained molecule has an additional...
stemming from misalignments in the structure, which can then be corrected by the designer.
Geometric models of nucleic acids for DNA nanotechnology
DNA nanotechnology
DNA nanotechnology is a branch of nanotechnology which uses the molecular recognition properties of DNA and other nucleic acids to create designed, artificial structures out of DNA for technological purposes. In this field, DNA is used as a structural material rather than as a carrier of genetic...
generally use reduced representations of the nucleic acid, because simulating every atom would be very computationally expensive for such large systems. Models with three pseudo-atoms per base pair, representing the two backbone sugars and the helix axis, have been reported to have a sufficient level of detail to predict experimental results. However, models with five pseudo-atoms per base pair, explicitly including the backbone phosphates, are also used.
Software for geometrical modeling of nucleic acids includes GIDEON,
Tiamat,
Nanoengineer-1,
and UNIQUIMER 3D.
Geometrical concerns are especially of interest in the design of DNA origami
DNA origami
DNA origami is the nanoscale folding of DNA to create arbitrary two and three dimensional shapes at the nanoscale. The specificity of the interactions between complementary base pairs make DNA a useful construction material through design of its base sequences...
, because the sequence is predetermined by the choice of scaffold strand. Software specifically for DNA origami design has been made, including caDNAno
and SARSE.
Applications
Nucleic acid design is used in DNA nanotechnologyDNA nanotechnology
DNA nanotechnology is a branch of nanotechnology which uses the molecular recognition properties of DNA and other nucleic acids to create designed, artificial structures out of DNA for technological purposes. In this field, DNA is used as a structural material rather than as a carrier of genetic...
to design strands which will self-assemble into a desired target structure. These include examples such as DNA machine
DNA machine
A DNA machine is a molecular machine constructed from DNA. Research into DNA machines was pioneered in the late 1980s by Nadrian Seeman and co-workers from New York University...
s, periodic two- and three-dimensional lattices, polyhedra, and DNA origami
DNA origami
DNA origami is the nanoscale folding of DNA to create arbitrary two and three dimensional shapes at the nanoscale. The specificity of the interactions between complementary base pairs make DNA a useful construction material through design of its base sequences...
. It can also be used to create sets of nucleic acid strands which are "orthogonal", or non-interacting with each other, so as to minimize or eliminate spurious interactions. This is useful in DNA computing
DNA computing
DNA computing is a form of computing which uses DNA, biochemistry and molecular biology, instead of the traditional silicon-based computer technologies. DNA computing, or, more generally, biomolecular computing, is a fast developing interdisciplinary area...
, as well as for molecular barcoding applications in chemical biology
Chemical biology
Chemical biology is a scientific discipline spanning the fields of chemistry and biology that involves the application of chemical techniques and tools, often compounds produced through synthetic chemistry, to the study and manipulation of biological systems. This is a subtle difference from...
and biotechnology
Biotechnology
Biotechnology is a field of applied biology that involves the use of living organisms and bioprocesses in engineering, technology, medicine and other fields requiring bioproducts. Biotechnology also utilizes these products for manufacturing purpose...
.
Further reading
—A review of approaches to nucleic acid primary structure design.—A comparison and evaluation of a number of heuristic and thermodynamic methods for nucleic acid design.
—One of the earliest papers on nucleic acid design, describing the use of sequence symmetry minimization to construct immoble branched junctions.
—A review comparing the capabilities of available nucleic acid design software.

