
Nucleic acid secondary structure
Encyclopedia
The secondary structure of a nucleic acid molecule refers to the basepairing interactions within a single molecule or set of interacting molecules, and can be represented as a list of bases which are paired in a nucleic acid molecule.
The secondary structures of biological DNA
's and RNA
's tend to be different: biological DNA mostly exists as fully base pair
ed double helices, while biological RNA is single stranded and often forms complicated base-pairing interactions due to its increased ability to form hydrogen bond
s stemming from the extra hydroxyl
group in the ribose
sugar.
In a non-biological context, secondary structure is a vital consideration in the rational design
of nucleic acid structures for DNA nanotechnology
and DNA computing
, since the pattern of basepairing ultimately determines the overall structure of the molecules.
, two nucleotide
s on opposite complementary
DNA
or RNA
strands that are connected via hydrogen bond
s are called a base pair (often abbreviated bp). In the canonical Watson-Crick base pairing, adenine
(A) forms a base pair with thymine
(T), as and guanine
(G) with cytosine
(C) in DNA. In RNA, thymine
is replaced by uracil
(U). Alternate hydrogen bonding patterns, such as the wobble base pair
and Hoogsteen base pair
, also occur—particularly in RNA—giving rise to complex and functional tertiary structures
. Importantly, pairing is the mechanism by which codons on messenger RNA
molecules are recognized by anticodons on transfer RNA
during protein translation
. Some DNA- or RNA-binding enzymes can recognize specific base pairing patterns that identify particular regulatory regions of genes.
Hydrogen bond
ing is the chemical mechanism that underlies the base-pairing rules described above. Appropriate geometrical correspondence of hydrogen bond donors and acceptors allows only the "right" pairs to form stably. DNA with high GC-content is more stable than DNA with low GC-content
, but contrary to popular belief, the hydrogen bonds do not stabilize the DNA significantly and stabilization is mainly due to stacking
interactions.
The larger nucleobase
s, adenine and guanine, are members of a class of doubly ringed chemical structures called purine
s; the smaller nucleobases, cytosine and thymine (and uracil), are members of a class of singly ringed chemical structures called pyrimidine
s. Purines are only complementary with pyrimidines: pyrimidine-pyrimidine pairings are energetically unfavorable because the molecules are too far apart for hydrogen bonding to be established; purine-purine pairings are energetically unfavorable because the molecules are too close, leading to overlap repulsion. The only other possible pairings are GT and AC; these pairings are mismatches because the pattern of hydrogen donors and acceptors do not correspond. The GU wobble base pair
, with two hydrogen bonds, does occur fairly often in RNA
.
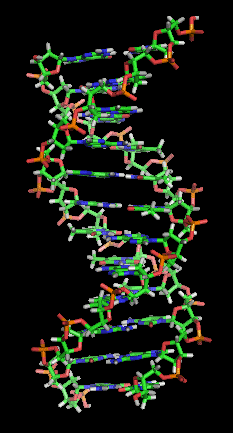
Hybridization is the process of complementary
base pair
s binding to form a double helix. Melting is the process by which the interactions between the strands of the double helix are broken, separating the two nucleic acid strands. These bonds are weak, easily separated by gentle heating, enzyme
s, or physical force. Melting occurs preferentially at certain points in the nucleic acid. T and A rich sequences are more easily melted than C and G rich regions. Particular base steps are also susceptible to DNA melting, particularly T A and T G base steps. These mechanical features are reflected by the use of sequences such as TATAA
at the start of many genes to assist RNA polymerase in melting the DNA for transcription.
Strand separation by gentle heating, as used in PCR, is simple providing the molecules have fewer than about 10,000 base pairs (10 kilobase pairs, or 10 kbp). The intertwining of the DNA strands makes long segments difficult to separate. The cell avoids this problem by allowing its DNA-melting enzymes (helicase
s) to work concurrently with topoisomerase
s, which can chemically cleave the phosphate backbone of one of the strands so that it can swivel around the other. Helicase
s unwind the strands to facilitate the advance of sequence-reading enzymes such as DNA polymerase
.
s, pseudoknot
s, and stem-loop
s.
in nuleic acid molecules which is intimately connected with the molecule's secondary structure. A double helix is formed by regions of many consecutive base pairs.
The DNA double helix is a right-handed spiral polymer of nucleic acids, held together by nucleotides which base pair together. A single turn of the helix constitutes about ten nucleotides, and contains a major groove and minor groove, the major groove being wider than the minor groove. Given the difference in widths of the major groove and minor groove, many proteins which bind to DNA do so through the wider major groove. Many double-helical forms are possible; for DNA the three biologically relevant forms are A-DNA
, B-DNA, and Z-DNA
, while RNA double helices have structures similar to the A form of DNA.
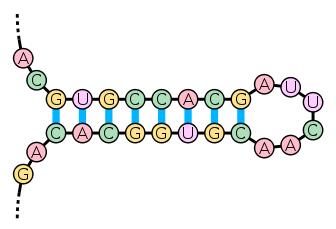
The secondary structure of nucleic acid molecules can often be uniquely decomposed into stems and loops. The stem-loop
structure in which a base-paired helix ends in a short unpaired loop is extremely common and is a building block for larger structural motifs such as cloverleaf structures, which are four-helix junctions such as those found in transfer RNA
. Internal loops (a short series of unpaired bases in a longer paired helix) and bulges (regions in which one strand of a helix has "extra" inserted bases with no counterparts in the opposite strand) are also frequent.
There are many secondary structure elements of functional importance to biological RNA's; some famous examples are the Rho-independent terminator
stem-loops and the tRNA cloverleaf
. There is a minor industry of researchers attempting to determine the secondary structure of RNA molecules. Approaches include both experimental and computational
methods (see also the List of RNA structure prediction software).
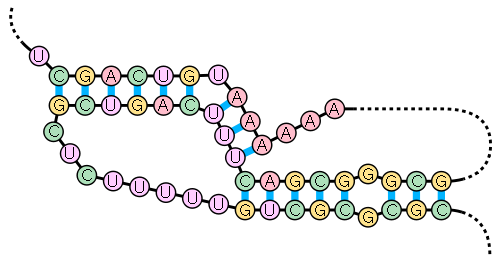 A pseudoknot is an RNA secondary structure containing at least two stem-loop
A pseudoknot is an RNA secondary structure containing at least two stem-loop
structures in which half of one stem is intercalated between the two halves of another stem. Pseudoknots fold into knot-shaped three-dimensional conformations but are not true topological knots
. The base pair
ing in pseudoknots is not well nested; that is, base pairs occur that "overlap" one another in sequence position. This makes the presence of general pseudoknots in RNA sequences impossible to predict
by the standard method of dynamic programming
, which uses a recursive scoring system to identify paired stems and consequently cannot detect non-nested base pairs with the most common algorithms. Limited subclasses of pseudoknots can be predicted using dynamic programs described in.
Newer structure prediction techniques such as stochastic context-free grammar
s also do not take pseudoknots into account.
Several important biological processes rely on RNA molecules that form pseudoknots. For example, the RNA component of human telomerase
contains a pseudoknot that is critical for activity. Though DNA can also form pseudoknots, they are generally not present in biological DNA.
uses predicted RNA secondary structures in searching a genome
for noncoding but functional forms of RNA. For example, microRNA
s have canonical long stem-loop structures interrupted by small internal loops. A general method of calculating probable RNA secondary structure is dynamic programming
, although this has the disadvantage that it cannot detect pseudoknot
s or other cases in which base pairs are not fully nested. More general methods are based on stochastic context-free grammar
s. A web server that implements a type of dynamic programming is Mfold.
For many RNA molecules, the secondary structure is highly important to the correct function of the RNA — often more so than the actual sequence. This fact aids in the analysis of non-coding RNA
sometimes termed "RNA genes". RNA secondary structure can be predicted with some accuracy by computer and many bioinformatics
applications use some notion of secondary structure in analysis of RNA.
The secondary structures of biological DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
's and RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
's tend to be different: biological DNA mostly exists as fully base pair
Base pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
ed double helices, while biological RNA is single stranded and often forms complicated base-pairing interactions due to its increased ability to form hydrogen bond
Hydrogen bond
A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond...
s stemming from the extra hydroxyl
Hydroxyl
A hydroxyl is a chemical group containing an oxygen atom covalently bonded with a hydrogen atom. In inorganic chemistry, the hydroxyl group is known as the hydroxide ion, and scientists and reference works generally use these different terms though they refer to the same chemical structure in...
group in the ribose
Ribose
Ribose is an organic compound with the formula C5H10O5; specifically, a monosaccharide with linear form H––4–H, which has all the hydroxyl groups on the same side in the Fischer projection....
sugar.
In a non-biological context, secondary structure is a vital consideration in the rational design
Nucleic acid design
Nucleic acid design is the process of generating a set of nucleic acid base sequences that will associate into a desired conformation. Nucleic acid design is central to the fields of DNA nanotechnology and DNA computing...
of nucleic acid structures for DNA nanotechnology
DNA nanotechnology
DNA nanotechnology is a branch of nanotechnology which uses the molecular recognition properties of DNA and other nucleic acids to create designed, artificial structures out of DNA for technological purposes. In this field, DNA is used as a structural material rather than as a carrier of genetic...
and DNA computing
DNA computing
DNA computing is a form of computing which uses DNA, biochemistry and molecular biology, instead of the traditional silicon-based computer technologies. DNA computing, or, more generally, biomolecular computing, is a fast developing interdisciplinary area...
, since the pattern of basepairing ultimately determines the overall structure of the molecules.
Base pairing
In molecular biologyMolecular biology
Molecular biology is the branch of biology that deals with the molecular basis of biological activity. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry...
, two nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
s on opposite complementary
Complementarity (molecular biology)
In molecular biology, complementarity is a property of double-stranded nucleic acids such as DNA, as well as DNA:RNA duplexes. Each strand is complementary to the other in that the base pairs between them are non-covalently connected via two or three hydrogen bonds...
DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
or RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
strands that are connected via hydrogen bond
Hydrogen bond
A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond...
s are called a base pair (often abbreviated bp). In the canonical Watson-Crick base pairing, adenine
Adenine
Adenine is a nucleobase with a variety of roles in biochemistry including cellular respiration, in the form of both the energy-rich adenosine triphosphate and the cofactors nicotinamide adenine dinucleotide and flavin adenine dinucleotide , and protein synthesis, as a chemical component of DNA...
(A) forms a base pair with thymine
Thymine
Thymine is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine nucleobase. As the name suggests, thymine may be derived by methylation of uracil at...
(T), as and guanine
Guanine
Guanine is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine . In DNA, guanine is paired with cytosine. With the formula C5H5N5O, guanine is a derivative of purine, consisting of a fused pyrimidine-imidazole ring system with...
(G) with cytosine
Cytosine
Cytosine is one of the four main bases found in DNA and RNA, along with adenine, guanine, and thymine . It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached . The nucleoside of cytosine is cytidine...
(C) in DNA. In RNA, thymine
Thymine
Thymine is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine nucleobase. As the name suggests, thymine may be derived by methylation of uracil at...
is replaced by uracil
Uracil
Uracil is one of the four nucleobases in the nucleic acid of RNA that are represented by the letters A, G, C and U. The others are adenine, cytosine, and guanine. In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine.Uracil is a common and...
(U). Alternate hydrogen bonding patterns, such as the wobble base pair
Wobble base pair
In molecular biology, a wobble base pair is a non-Watson-Crick base pairing between two nucleotides in RNA molecules. The four main wobble base pairs are guanine-uracil, inosine-uracil, inosine-adenine, and inosine-cytosine . The thermodynamic stability of a wobble base pair is comparable to that...
and Hoogsteen base pair
Hoogsteen base pair
A Hoogsteen base pair is a variation of base-pairing in nucleic acids such as the A•T pair. In this manner, two nucleobases on each strand can be held together by hydrogen bonds in the major groove...
, also occur—particularly in RNA—giving rise to complex and functional tertiary structures
Nucleic acid tertiary structure
300px|thumb|upright|alt = Colored dice with checkered background|Example of a large catalytic RNA. The self-splicing group II intron from Oceanobacillus iheyensis....
. Importantly, pairing is the mechanism by which codons on messenger RNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
molecules are recognized by anticodons on transfer RNA
Transfer RNA
Transfer RNA is an adaptor molecule composed of RNA, typically 73 to 93 nucleotides in length, that is used in biology to bridge the three-letter genetic code in messenger RNA with the twenty-letter code of amino acids in proteins. The role of tRNA as an adaptor is best understood by...
during protein translation
Translation (genetics)
In molecular biology and genetics, translation is the third stage of protein biosynthesis . In translation, messenger RNA produced by transcription is decoded by the ribosome to produce a specific amino acid chain, or polypeptide, that will later fold into an active protein...
. Some DNA- or RNA-binding enzymes can recognize specific base pairing patterns that identify particular regulatory regions of genes.
Hydrogen bond
Hydrogen bond
A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond...
ing is the chemical mechanism that underlies the base-pairing rules described above. Appropriate geometrical correspondence of hydrogen bond donors and acceptors allows only the "right" pairs to form stably. DNA with high GC-content is more stable than DNA with low GC-content
GC-content
In molecular biology and genetics, GC-content is the percentage of nitrogenous bases on a DNA molecule that are either guanine or cytosine . This may refer to a specific fragment of DNA or RNA, or that of the whole genome...
, but contrary to popular belief, the hydrogen bonds do not stabilize the DNA significantly and stabilization is mainly due to stacking
Stacking (chemistry)
In chemistry, pi stacking refers to attractive, noncovalent interactions between aromatic rings. These interactions are historically thought to be important in to base stacking of DNA nucleotides, protein folding, template-directed synthesis, materials science, and molecular recognition, although...
interactions.
The larger nucleobase
Nucleobase
Nucleobases are a group of nitrogen-based molecules that are required to form nucleotides, the basic building blocks of DNA and RNA. Nucleobases provide the molecular structure necessary for the hydrogen bonding of complementary DNA and RNA strands, and are key components in the formation of stable...
s, adenine and guanine, are members of a class of doubly ringed chemical structures called purine
Purine
A purine is a heterocyclic aromatic organic compound, consisting of a pyrimidine ring fused to an imidazole ring. Purines, including substituted purines and their tautomers, are the most widely distributed kind of nitrogen-containing heterocycle in nature....
s; the smaller nucleobases, cytosine and thymine (and uracil), are members of a class of singly ringed chemical structures called pyrimidine
Pyrimidine
Pyrimidine is a heterocyclic aromatic organic compound similar to benzene and pyridine, containing two nitrogen atoms at positions 1 and 3 of the six-member ring...
s. Purines are only complementary with pyrimidines: pyrimidine-pyrimidine pairings are energetically unfavorable because the molecules are too far apart for hydrogen bonding to be established; purine-purine pairings are energetically unfavorable because the molecules are too close, leading to overlap repulsion. The only other possible pairings are GT and AC; these pairings are mismatches because the pattern of hydrogen donors and acceptors do not correspond. The GU wobble base pair
Wobble base pair
In molecular biology, a wobble base pair is a non-Watson-Crick base pairing between two nucleotides in RNA molecules. The four main wobble base pairs are guanine-uracil, inosine-uracil, inosine-adenine, and inosine-cytosine . The thermodynamic stability of a wobble base pair is comparable to that...
, with two hydrogen bonds, does occur fairly often in RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
.
Nucleic acid hybridization

| Step | Melting ΔG /Kcal mol−1 |
|---|---|
| T A | -0.12 |
| T G or C A | -0.78 |
| C G | -1.44 |
| A G or C T | -1.29 |
| A A or T T | -1.04 |
| A T | -1.27 |
| G A or T C | -1.66 |
| C C or G G | -1.97 |
| A C or G T | -2.04 |
| G C | -2.70 |
Hybridization is the process of complementary
Complementarity (molecular biology)
In molecular biology, complementarity is a property of double-stranded nucleic acids such as DNA, as well as DNA:RNA duplexes. Each strand is complementary to the other in that the base pairs between them are non-covalently connected via two or three hydrogen bonds...
base pair
Base pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
s binding to form a double helix. Melting is the process by which the interactions between the strands of the double helix are broken, separating the two nucleic acid strands. These bonds are weak, easily separated by gentle heating, enzyme
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
s, or physical force. Melting occurs preferentially at certain points in the nucleic acid. T and A rich sequences are more easily melted than C and G rich regions. Particular base steps are also susceptible to DNA melting, particularly T A and T G base steps. These mechanical features are reflected by the use of sequences such as TATAA
TATA box
The TATA box is a DNA sequence found in the promoter region of genes in archaea and eukaryotes; approximately 24% of human genes contain a TATA box within the core promoter....
at the start of many genes to assist RNA polymerase in melting the DNA for transcription.
Strand separation by gentle heating, as used in PCR, is simple providing the molecules have fewer than about 10,000 base pairs (10 kilobase pairs, or 10 kbp). The intertwining of the DNA strands makes long segments difficult to separate. The cell avoids this problem by allowing its DNA-melting enzymes (helicase
Helicase
Helicases are a class of enzymes vital to all living organisms. They are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two annealed nucleic acid strands using energy derived from ATP hydrolysis.-Function:Many cellular processes Helicases are a...
s) to work concurrently with topoisomerase
Topoisomerase
Topoisomerases are enzymes that regulate the overwinding or underwinding of DNA. The winding problem of DNA arises due to the intertwined nature of its double helical structure. For example, during DNA replication, DNA becomes overwound ahead of a replication fork...
s, which can chemically cleave the phosphate backbone of one of the strands so that it can swivel around the other. Helicase
Helicase
Helicases are a class of enzymes vital to all living organisms. They are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two annealed nucleic acid strands using energy derived from ATP hydrolysis.-Function:Many cellular processes Helicases are a...
s unwind the strands to facilitate the advance of sequence-reading enzymes such as DNA polymerase
DNA polymerase
A DNA polymerase is an enzyme that helps catalyze in the polymerization of deoxyribonucleotides into a DNA strand. DNA polymerases are best known for their feedback role in DNA replication, in which the polymerase "reads" an intact DNA strand as a template and uses it to synthesize the new strand....
.
Secondary structure motifs
Nucleic acid secondary structure is generally divided into helices (contiguous base pairs), and various kinds of loops (unpaired nucleotides surrounded by helices). Frequently these elements, or combinations of them, can be further classified, for example, tetraloopTetraloop
Tetraloops are a type of four-base hairpin loop motifs in RNA secondary structure that cap many double helices. Three types of tetraloops are common in ribosomal RNA: GNRA, UNCG and CUUG. The GNRA tetraloop has a guanine-adenine base-pair where the guanine is 5' to the helix and the adenine is 3'...
s, pseudoknot
Pseudoknot
A pseudoknot is a nucleic acid secondary structure containing at least two stem-loop structures in which half of one stem is intercalated between the two halves of another stem. The pseudoknot was first recognized in the turnip yellow mosaic virus in 1982...
s, and stem-loop
Stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions,...
s.
Double helix
The double helix is an important tertiary structureNucleic acid tertiary structure
300px|thumb|upright|alt = Colored dice with checkered background|Example of a large catalytic RNA. The self-splicing group II intron from Oceanobacillus iheyensis....
in nuleic acid molecules which is intimately connected with the molecule's secondary structure. A double helix is formed by regions of many consecutive base pairs.
The DNA double helix is a right-handed spiral polymer of nucleic acids, held together by nucleotides which base pair together. A single turn of the helix constitutes about ten nucleotides, and contains a major groove and minor groove, the major groove being wider than the minor groove. Given the difference in widths of the major groove and minor groove, many proteins which bind to DNA do so through the wider major groove. Many double-helical forms are possible; for DNA the three biologically relevant forms are A-DNA
A-DNA
A-DNA is one of the many possible double helical structures of DNA. A-DNA is thought to be one of three biologically active double helical structures along with B- and Z-DNA. It is a right-handed double helix fairly similar to the more common and well-known B-DNA form, but with a shorter more...
, B-DNA, and Z-DNA
Z-DNA
Z-DNA is one of the many possible double helical structures of DNA. It is a left-handed double helical structure in which the double helix winds to the left in a zig-zag pattern...
, while RNA double helices have structures similar to the A form of DNA.
Stem-loop structures

The secondary structure of nucleic acid molecules can often be uniquely decomposed into stems and loops. The stem-loop
Stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions,...
structure in which a base-paired helix ends in a short unpaired loop is extremely common and is a building block for larger structural motifs such as cloverleaf structures, which are four-helix junctions such as those found in transfer RNA
Transfer RNA
Transfer RNA is an adaptor molecule composed of RNA, typically 73 to 93 nucleotides in length, that is used in biology to bridge the three-letter genetic code in messenger RNA with the twenty-letter code of amino acids in proteins. The role of tRNA as an adaptor is best understood by...
. Internal loops (a short series of unpaired bases in a longer paired helix) and bulges (regions in which one strand of a helix has "extra" inserted bases with no counterparts in the opposite strand) are also frequent.
There are many secondary structure elements of functional importance to biological RNA's; some famous examples are the Rho-independent terminator
Intrinsic termination
Intrinsic termination is a mechanism in prokaryotes that causes mRNA transcription to be stopped. In this mechanism, the mRNA contains a sequence that can base pair with itself to form a stem-loop structure 7-20 base pairs in length that is also rich in cytosine-guanine base pairs...
stem-loops and the tRNA cloverleaf
Transfer RNA
Transfer RNA is an adaptor molecule composed of RNA, typically 73 to 93 nucleotides in length, that is used in biology to bridge the three-letter genetic code in messenger RNA with the twenty-letter code of amino acids in proteins. The role of tRNA as an adaptor is best understood by...
. There is a minor industry of researchers attempting to determine the secondary structure of RNA molecules. Approaches include both experimental and computational
Nucleic acid structure prediction
Nucleic acid structure prediction is a computational method to determine nucleic acid secondary and tertiary structure from its sequence. Secondary structure can be predicted from a single or from several nucleic acid sequences...
methods (see also the List of RNA structure prediction software).
Pseudoknots

Stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions,...
structures in which half of one stem is intercalated between the two halves of another stem. Pseudoknots fold into knot-shaped three-dimensional conformations but are not true topological knots
Knot (mathematics)
In mathematics, a knot is an embedding of a circle in 3-dimensional Euclidean space, R3, considered up to continuous deformations . A crucial difference between the standard mathematical and conventional notions of a knot is that mathematical knots are closed—there are no ends to tie or untie on a...
. The base pair
Base pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
ing in pseudoknots is not well nested; that is, base pairs occur that "overlap" one another in sequence position. This makes the presence of general pseudoknots in RNA sequences impossible to predict
Secondary structure prediction
Secondary structure prediction is a set of techniques in bioinformatics that aim to predict the secondary structures of proteins and nucleic acid sequences based only on knowledge of their primary structure...
by the standard method of dynamic programming
Dynamic programming
In mathematics and computer science, dynamic programming is a method for solving complex problems by breaking them down into simpler subproblems. It is applicable to problems exhibiting the properties of overlapping subproblems which are only slightly smaller and optimal substructure...
, which uses a recursive scoring system to identify paired stems and consequently cannot detect non-nested base pairs with the most common algorithms. Limited subclasses of pseudoknots can be predicted using dynamic programs described in.
Newer structure prediction techniques such as stochastic context-free grammar
Stochastic context-free grammar
A stochastic context-free grammar is a context-free grammar in which each production is augmented with a probability...
s also do not take pseudoknots into account.
Several important biological processes rely on RNA molecules that form pseudoknots. For example, the RNA component of human telomerase
Telomerase
Telomerase is an enzyme that adds DNA sequence repeats to the 3' end of DNA strands in the telomere regions, which are found at the ends of eukaryotic chromosomes. This region of repeated nucleotide called telomeres contains non-coding DNA material and prevents constant loss of important DNA from...
contains a pseudoknot that is critical for activity. Though DNA can also form pseudoknots, they are generally not present in biological DNA.
Secondary structure prediction
One application of bioinformaticsBioinformatics
Bioinformatics is the application of computer science and information technology to the field of biology and medicine. Bioinformatics deals with algorithms, databases and information systems, web technologies, artificial intelligence and soft computing, information and computation theory, software...
uses predicted RNA secondary structures in searching a genome
Genome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
for noncoding but functional forms of RNA. For example, microRNA
Mirna
Mirna may refer to:geographical entities* Mirna , a river in Istria, Croatia* Mirna , a river in Slovenia, tributary of the river Sava* Mirna , a settlement in the municipality of Mirna in Southeastern Sloveniapeople...
s have canonical long stem-loop structures interrupted by small internal loops. A general method of calculating probable RNA secondary structure is dynamic programming
Dynamic programming
In mathematics and computer science, dynamic programming is a method for solving complex problems by breaking them down into simpler subproblems. It is applicable to problems exhibiting the properties of overlapping subproblems which are only slightly smaller and optimal substructure...
, although this has the disadvantage that it cannot detect pseudoknot
Pseudoknot
A pseudoknot is a nucleic acid secondary structure containing at least two stem-loop structures in which half of one stem is intercalated between the two halves of another stem. The pseudoknot was first recognized in the turnip yellow mosaic virus in 1982...
s or other cases in which base pairs are not fully nested. More general methods are based on stochastic context-free grammar
Stochastic context-free grammar
A stochastic context-free grammar is a context-free grammar in which each production is augmented with a probability...
s. A web server that implements a type of dynamic programming is Mfold.
For many RNA molecules, the secondary structure is highly important to the correct function of the RNA — often more so than the actual sequence. This fact aids in the analysis of non-coding RNA
Non-coding RNA
A non-coding RNA is a functional RNA molecule that is not translated into a protein. Less-frequently used synonyms are non-protein-coding RNA , non-messenger RNA and functional RNA . The term small RNA is often used for short bacterial ncRNAs...
sometimes termed "RNA genes". RNA secondary structure can be predicted with some accuracy by computer and many bioinformatics
Bioinformatics
Bioinformatics is the application of computer science and information technology to the field of biology and medicine. Bioinformatics deals with algorithms, databases and information systems, web technologies, artificial intelligence and soft computing, information and computation theory, software...
applications use some notion of secondary structure in analysis of RNA.
See also
- DNA nanotechnologyDNA nanotechnologyDNA nanotechnology is a branch of nanotechnology which uses the molecular recognition properties of DNA and other nucleic acids to create designed, artificial structures out of DNA for technological purposes. In this field, DNA is used as a structural material rather than as a carrier of genetic...
- Molecular models of DNAMolecular models of DNAMolecular models of DNA structures are representations of the molecular geometry and topology of Deoxyribonucleic acid molecules using one of several means, with the aim of simplifying and presenting the essential, physical and chemical, properties of DNA molecular structures either in vivo or in...
- DiProDBDiProDBDiProDB is a database designed to collect and analyse thermodynamic, structural and other dinucleotide properties....
. The database is designed to collect and analyse thermodynamic, structural and other dinucleotide properties.
External links
- MDDNA: Structural Bioinformatics of DNA
- Abalone — Commercial software for DNA modeling
- DNAlive: a web interface to compute DNA physical properties. Also allows cross-linking of the results with the UCSC Genome browserGenome browserA genome browser is a graphical interface for display of information from a biological database for genomic data. Genome browsers enable researchers to visualize and browse entire genomes with annotated data including gene prediction and structure, proteins, expression, regulation, variation,...
and DNA dynamics.

