
Two-hybrid screening
Encyclopedia
Molecular biology
Molecular biology is the branch of biology that deals with the molecular basis of biological activity. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry...
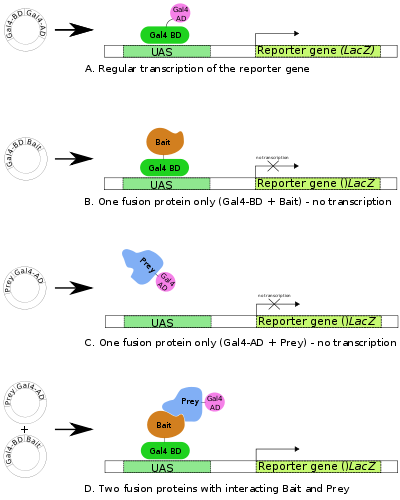
technique used to discover protein–protein interactions and protein–DNA interactions by testing for physical interactions (such as binding) between two protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s or a single protein and a DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
molecule, respectively.
The premise behind the test is the activation of downstream reporter gene
Reporter gene
In molecular biology, a reporter gene is a gene that researchers attach to a regulatory sequence of another gene of interest in cell culture, animals or plants. Certain genes are chosen as reporters because the characteristics they confer on organisms expressing them are easily identified and...
(s) by the binding of a transcription factor
Transcription factor
In molecular biology and genetics, a transcription factor is a protein that binds to specific DNA sequences, thereby controlling the flow of genetic information from DNA to mRNA...
onto an upstream activating sequence (UAS). For two-hybrid screening, the transcription factor is split into two separate fragments, called the binding domain (BD) and activating domain (AD). The BD is the domain
Protein domain
A protein domain is a part of protein sequence and structure that can evolve, function, and exist independently of the rest of the protein chain. Each domain forms a compact three-dimensional structure and often can be independently stable and folded. Many proteins consist of several structural...
responsible for binding
DNA-binding protein
DNA-binding proteins are proteins that are composed of DNA-binding domains and thus have a specific or general affinity for either single or double stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, because it exposes more functional groups that...
to the UAS and the AD is the domain responsible for the activation of transcription
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
.
History
Pioneered by Stanley Fields and Song in 1989, the technique was originally designed to detect protein–protein interactions using the GAL4 transcriptional activator of the yeast Saccharomyces cerevisiaeSaccharomyces cerevisiae
Saccharomyces cerevisiae is a species of yeast. It is perhaps the most useful yeast, having been instrumental to baking and brewing since ancient times. It is believed that it was originally isolated from the skin of grapes...
. The GAL4 protein activated transcription of a protein involved in galactose utilization, which formed the basis of selection. Since then, the same principle has been adapted to describe many alternative methods including some that detect protein–DNA interactions, DNA-DNA interactions and use Escherichia coli
Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
instead of yeast.
Basic premise
The key to the two-hybrid screen is that in most eukaryotic transcription factors, the activating and binding domains are modular and can function in close proximity to each other without direct binding. This means that even though the transcription factor is split into two fragments, it can still activate transcription when the two fragments are indirectly connected.The most common screening approach is the yeast two-hybrid assay. This system often utilizes a genetically engineered
Genetic engineering
Genetic engineering, also called genetic modification, is the direct human manipulation of an organism's genome using modern DNA technology. It involves the introduction of foreign DNA or synthetic genes into the organism of interest...
strain of yeast in which the biosynthesis
Biosynthesis
Biosynthesis is an enzyme-catalyzed process in cells of living organisms by which substrates are converted to more complex products. The biosynthesis process often consists of several enzymatic steps in which the product of one step is used as substrate in the following step...
of certain nutrients (usually amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
s or nucleic acid
Nucleic acid
Nucleic acids are biological molecules essential for life, and include DNA and RNA . Together with proteins, nucleic acids make up the most important macromolecules; each is found in abundance in all living things, where they function in encoding, transmitting and expressing genetic information...
s) is lacking. When grown on media that lacks these nutrients, the yeast fail to survive. This mutant yeast strain can be made to incorporate foreign DNA in the form of plasmid
Plasmid
In microbiology and genetics, a plasmid is a DNA molecule that is separate from, and can replicate independently of, the chromosomal DNA. They are double-stranded and, in many cases, circular...
s. In yeast two-hybrid screening, separate bait and prey plasmids are simultaneously introduced into the mutant yeast strain.
Plasmids are engineered to produce a protein product in which the DNA-binding domain (BD) fragment is fused onto a protein while another plasmid is engineered to produce a protein product in which the activation domain (AD) fragment is fused onto another protein. The protein fused to the BD may be referred to as the bait protein, and is typically a known protein the investigator is using to identify new binding partners. The protein fused to the AD may be referred to as the prey protein and can be either a single known protein or a library
Library (biology)
In molecular biology, a library is a collection of DNA fragments that is stored and propagated in a population of micro-organisms through the process of molecular cloning...
of known or unknown proteins. In this context, a library may consist of a collection of protein-encoding sequences that represent all the proteins expressed in a particular organism or tissue, or may be generated by synthesising random DNA sequences. Regardless of the source, they are subsequently incorporated into the protein-encoding sequence of a plasmid, which is then transfected into the cells chosen for the screening method. This technique, when using a library, assumes that each cell is transfected with no more than a single plasmid and that, therefore, each cell ultimately expresses no more than a single member from the protein library.
If the bait and prey proteins interact (i.e., bind), then the AD and BD of the transcription factor are indirectly connected, bringing the AD in proximity to the transcription start site and transcription of reporter gene(s) can occur. If the two proteins do not interact, there is no transcription of the reporter gene. In this way, a successful interaction between the fused protein is linked to a change in the cell phenotype.
The challenge of separating cells that express proteins that happen to interact with their counterpart fusion proteins from those that do not, is addressed in the following section.
Fixed domains
In any study, some of the protein domains, those under investigation, will be varied according to the goals of the study whereas other domains, those that are not themselves being investigated, will be kept constant. For example in a two-hybrid study to select DNA-binding domains, the DNA-binding domain, BD, will be varied whilst the two interacting proteins, the bait and prey, must be kept constant to maintain a strong binding between the BD and AD. There are a number of domains from which to choose the BD, bait and prey and AD, if these are to remain constant. In protein–protein interaction investigations, the BD may be chosen from any of many strong DNA-binding domains such as Zif268Zif268
EGR-1 also known as Zif268 or NGFI-A is a protein that in humans is encoded by the EGR1 gene....
. A frequent choice of bait and prey domains are residues 263–352 of yeast Gal11P with a N342V mutation and residues 58–97 of yeast Gal4, respectively. These domains can be used in both yeast- and bacterial-based selection techniques and are known to bind together strongly.
The AD chosen must be able to activate transcription of the reporter gene, using the cell's own transcription machinery. Thus, the variety of ADs available for use in yeast-based techniques may not be suited to use in their bacterial-based analogues. The herpes simplex virus-derived AD, VP16 and yeast Gal4 AD have been used with success in yeast whilst a portion of the α-subunit of E. coli RNA polymerase has been utilised in E. coli-based methods.
Whilst powerfully activating domains may allow greater sensitivity towards weaker interactions, conversely, a weaker AD may provide greater stringency.
Construction of expression plasmids
A number of engineered genetic sequences must be incorporated into the host cell to perform two-hybrid analysis or one of its derivative techniques. The considerations and methods used in the construction and delivery of these sequences differ according to the needs of the assay and the organism chosen as the experimental background.There are two broad categories of hybrid library: random libraries and cDNA-based libraries. A cDNA library
CDNA library
A cDNA library is a combination of cloned cDNA fragments inserted into a collection of host cells, which together constitute some portion of the transcriptome of the organism. cDNA is produced from fully transcribed mRNA found in the nucleus and therefore contains only the expressed genes of an...
is constituted by the cDNA produced through reverse transcription of mRNA collected from specific cells of types of cell. This library can be ligated into a construct so that it is attached to the BD or AD being used in the assay. A random library uses lengths of DNA of random sequence in place of these cDNA sections. A number of methods exist for the production of these random sequences, including cassette mutagenesis. Regardless of the source of the DNA library, it is ligated
DNA ligase
In molecular biology, DNA ligase is a specific type of enzyme, a ligase, that repairs single-stranded discontinuities in double stranded DNA molecules, in simple words strands that have double-strand break . Purified DNA ligase is used in gene cloning to join DNA molecules together...
into the appropriate place in the relevant plasmid/phagemid using the appropriate restriction endonucleases.
E. coli-specific considerations
By placing the hybrid proteins under the control of IPTGIPTG
Isopropyl β-D-1-thiogalactopyranoside, abbreviated IPTG, is a molecular biology reagent.This compound is used as a molecular mimic of allolactose, a lactose metabolite that triggers transcription of the lac operon...
-inducible lac promoters, they are expressed only on media supplemented with IPTG. Further, by including different antibiotic resistance genes in each genetic construct, the growth of non-transformed cells is easily prevented through culture on media containing the corresponding antibiotics. This is particularly important for counter selection methods in which a lack of interaction is needed for cell survival.
The reporter gene may be inserted into the E. coli genome by first inserting it into an episome, a type of plasmid with the ability to incorporate itself into the bacterial cell genome with a copy number of approximately one per cell.
The hybrid expression phagemids can be electroporated into E. coli XL-1 Blue cells which after amplification and infection with VCS-M13 helper phage, will yield a stock of library phage. These phage will each contain one single-stranded member of the phagemid library.
Recovery of protein information
Once the selection has been performed, the primary structurePrimary structure
The primary structure of peptides and proteins refers to the linear sequence of its amino acid structural units. The term "primary structure" was first coined by Linderstrøm-Lang in 1951...
of the proteins which display the appropriate characteristics must be determined. This is achieved by retrieval of the protein-encoding sequences (as originally inserted) from the cells showing the appropriate phenotype.
E. coli
The phagemid used to transform E. coli cells may be "rescued" from the selected cells by infecting them with VCS-M13 helper phage. The resulting phage particles that are produced contain the single-stranded phagemids and are used to infect XL-1 Blue cells. The double-stranded phagemids are subsequently collected from these XL-1 Blue cells, essentially reversing the process used to produce the original library phage. Finally, the DNA sequences are determined through dideoxy sequencing.Controlling sensitivity
The Escherichia coli-derived Tet-R repressor can be used in line with a conventional reporter gene and can be controlled by tetracycline or doxicycline (Tet-R inhibitors). Thus the expression of Tet-R is controlled by the standard two-hybrid system but the Tet-R in turn controls (represses) the expression of a previously mentioned reporter such as HIS3, through its Tet-R promoter. Tetracycline or its derivatives can then be used to regulate the sensitivity of a system utilising Tet-R.Sensitivity may also be controlled by varying the dependency of the cells on their reporter genes. For example, this effected by altering the concentration of histidine in the growth medium for his3-dependent cells and altering the concentration of streptomycin for aadA dependent cells. Selection-gene-dependency may also be controlled by applying an inhibitor of the selection gene at a suitable concentration. 3-Amino-1,2,4-triazole
3-Amino-1,2,4-triazole
3-Amino-1,2,4-triazole is a heterocyclic organic compound that consists of a 1,2,4-triazole substituted with an amino group.3-AT is a competitive inhibitor of the product of the HIS3 gene, imidazoleglycerol-phosphate dehydratase...
(3-AT) for example, is a competitive inhibitor of the HIS3-gene product and may be used to titrate the minimum level of HIS3 expression required for growth on histidine-deficient media.
Sensitivity may also be modulated by varying the number of operator sequences in the reporter DNA.
Non-fusion proteins
A third, non-fusion protein may be co-expressed with two fusion proteins. Depending on the investigation, the third protein may modify one of the fusion proteins or mediate or interfere with their interaction.Co-expression of the third protein may be necessary for modification or activation of one or both of the fusion proteins. For example S. cerevisiae possesses no endogenous tyrosine kinase. If an investigation involves a protein that requires tyrosine phosphorylation, the kinase must be supplied in the form of a tyrosine kinase gene.
The non-fusion protein may mediate the interaction by binding both fusion proteins simultaneously, as in the case of ligand-dependent receptor dimerization.
For a protein with an interacting partner, its functional homology to other proteins may be assessed by supplying the third protein in non-fusion form, which then may or may not compete with the fusion-protein for its binding partner. Binding between the third protein and the other fusion protein will interrupt the formation of the reporter expression activation complex and thus reduce reporter expression, leading to the distinguishing change in phenotype.
Split-ubiquitin yeast two-hybrid
One limitation of classic yeast two-hybrid screens is that they are limited to soluble proteins. It is therefore impossible to use them to study the protein–protein interactions between insoluble integral membrane proteinIntegral membrane protein
An integral membrane protein is a protein molecule that is permanently attached to the biological membrane. Proteins that cross the membrane are surrounded by "annular" lipids, which are defined as lipids that are in direct contact with a membrane protein...
s. The split-ubiquitin system provides a method for overcoming this limitation. In the split-ubiquitin system, two integral membrane proteins to be studied are fused to two different ubiquitin
Ubiquitin
Ubiquitin is a small regulatory protein that has been found in almost all tissues of eukaryotic organisms. Among other functions, it directs protein recycling.Ubiquitin can be attached to proteins and label them for destruction...
moieties: a C-terminal ubiquitin moiety ("Cub", residues 35–76) and an N-terminal ubiquitin moiety ("Nub", residues 1–34). These fused proteins are called the bait and prey, respectively. In addition to being fused to an integral membrane protein, the Cub moiety is also fused to a transcription factor
Transcription factor
In molecular biology and genetics, a transcription factor is a protein that binds to specific DNA sequences, thereby controlling the flow of genetic information from DNA to mRNA...
(TF) that can be cleaved off by ubiquitin specific protease
Protease
A protease is any enzyme that conducts proteolysis, that is, begins protein catabolism by hydrolysis of the peptide bonds that link amino acids together in the polypeptide chain forming the protein....
s. Upon bait–prey interaction, Nub and Cub-moieties assemble, reconstituting the split-ubiquitin. The reconstituted split-ubiquitin molecule is recognized by ubiquitin specific proteases, which cleave off the reporter protein, allowing it to induce the transcription of reporter gene
Reporter gene
In molecular biology, a reporter gene is a gene that researchers attach to a regulatory sequence of another gene of interest in cell culture, animals or plants. Certain genes are chosen as reporters because the characteristics they confer on organisms expressing them are easily identified and...
s.
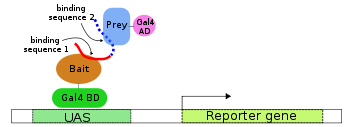
One-hybrid
The one-hybrid variation of this technique is designed to investigate protein–DNA interactions and uses a single fusion protein in which the AD is linked directly to the binding domain. The binding domain in this case however is not necessarily of fixed sequence as in two-hybrid protein–protein analysis but may be constituted by a library. This library can be selected against the desired target sequence, which is inserted in the promoter region of the reporter gene construct. In a positive-selection system, a binding domain that successfully binds the UAS and allows transcription is thus selected.Note that selection of DNA-binding domains is not necessarily performed using a one-hybrid system, but may also be performed using a two-hybrid system in which the binding domain is varied and the bait and prey proteins are kept constant.
Three-hybrid
One-two-hybrid
Simultaneous use of the one- and two-hybrid methods (that is, simultaneous protein–protein and protein–DNA interaction) is known as a one-two-hybrid approach and expected to increase the stringency of the screen.Host organism
Although theoretically, any living cell might be used as the background to a two-hybrid analysis, there are practical considerations that dictate which is chosen. The chosen cell line should be relatively cheap and easy to culture and sufficiently robust to withstand application of the investigative methods and reagents.Yeast
S. cerevisiae was the model organism used during the two-hybrid technique's inception. It has several characteristics that make it a robust organism to host the interaction, including the ability to form tertiary protein structures, neutral internal pH, enhanced ability to form disulfide bonds and reduced-state glutathione among other cytosolic buffer factors, to maintain a hospitable internal environment. The yeast model can be manipulated through non-molecular techniques and its complete genome sequence is known. Yeast systems are tolerant of diverse culture conditions and harsh chemicals that could not be applied to mammalian tissue cultures.Proteins from as small as eight to as large as 750 amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
s have been studied using yeast.
E. coli
E. coli-based methods have several characteristics that may make them preferable to yeast-based homologues. The higher transformation efficiency and faster rate of growth lends E. coli to the use of larger libraries (in excess of 108). A low false positive rate of approximately 3x10−8, the absence of requirement for a nuclear localisation signal to be included in the protein sequence and the ability to study proteins that would be toxic to yeast may also be major factors to consider when choosing an experimental background organism.It may be of note that the methylation activity of certain E. coli DNA methyltransferase
DNA methyltransferase
In biochemistry, the DNA methyltransferase family of enzymescatalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions...
proteins may interfere with some DNA-binding protein selections. If this is anticipated, the use of an E. coli strain that is defective for a particular methyltransferase may be an obvious solution.
Determination of sequences crucial for interaction
By changing specific amino acids by mutating the corresponding DNA base-pairs in the plasmids used, the importance of those amino acid residues in maintaining the interaction can be determined.After using bacterial cell-based method to select DNA-binding proteins, it is necessary to check the specificity of these domains as there is a limit to the extent to which the bacterial cell genome can act as a sink for domains with an affinity for other sequences (or indeed, a general affinity for DNA).
Drug and poison discovery
Protein–protein signalling interactions pose suitable therapeutic targets due to their specificity and pervasiveness. The random drug discovery approach uses compound banks that comprise random chemical structures, and requires a high-throughput method to test these structures in their intended target.The cell chosen for the investigation can be specifically engineered to mirror the molecular aspect that the investigator intends to study and then used to identify new human or animal therapeutics or anti-pest agents.
Determination of protein function
By determination of the interaction partners of unknown proteins, the possible functions of these new proteins may be inferred. This can be done using a single known protein against a library of unknown proteins or conversely, by selecting from a library of known proteins using a single protein of unknown function.Zinc finger protein selection
To select zinc finger proteins (ZFPs) for protein engineeringProtein engineering
Protein engineering is the process of developing useful or valuable proteins. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles....
, methods adapted from the two-hybrid screening technique have been used with success. A ZFP is itself a DNA-binding protein used in the construction of custom DNA-binding domains that bind to a desired DNA sequence.
By using a selection gene with the desired target sequence included in the UAS, and randomising the relevant amino acid sequences to produce a ZFP library, cells that host a DNA-ZFP interaction with the required characteristics can be selected. Each ZFP typically recognises only 3–4 base pairs, so to prevent recognition of sites outside the UAS, the randomised ZFP is engineered into a 'scaffold' consisting of another two ZFPs of constant sequence. The UAS is thus designed to include the target sequence of the constant scaffold in addition to the sequence for which a ZFP is selected.
A number of other DNA-binding domains may also be investigated using this system.
Strengths and weaknesses
Two-hybrid screens are now routinely performed in many labs. They can provide an important first hint for the identification of interaction partners. Moreover, the assay is scalable, which makes it possible to screen for interactions among many proteins.The main criticism applied to the yeast two-hybrid screen of protein–protein interactions is the possibility of a high number of false positive (and false negative) identifications. The exact rate of false positive results is not known, but estimates are as high as 70%. The reason for this high error rate lies in the principle of the screen: The assay investigates the interaction between (i) overexpressed (ii) fusion proteins in the (iii) yeast (iv) nucleus. Each of these points (i–iv) alone can give rise to false results. For example, overexpression can result in non-specific interactions. Moreover, a mammalian protein is sometimes not correctly modified in yeast (e.g., missing phosphorylation
Phosphorylation
Phosphorylation is the addition of a phosphate group to a protein or other organic molecule. Phosphorylation activates or deactivates many protein enzymes....
), which can also lead to false results. Finally, some proteins might specifically interact when they are co-expressed in the yeast, although in reality they are never present in the same cell at the same time. Due to the combined effects of all error sources the overall confidence of the yeast two-hybrid assay is rather low. However, yeast two-hybrid data is shown to be of similar quality to data generated by the alternative approach of coaffinity purification followed by mass spectrometry
Mass spectrometry
Mass spectrometry is an analytical technique that measures the mass-to-charge ratio of charged particles.It is used for determining masses of particles, for determining the elemental composition of a sample or molecule, and for elucidating the chemical structures of molecules, such as peptides and...
(AP/MS). The probability of generating false positives means that all interactions should be confirmed by a high confidence assay, for example co-immunoprecipitation of the endogenous proteins, which is difficult for large scale protein–protein interaction data.
See also
- Phage displayPhage displayPhage display is a method for the study of protein–protein, protein–peptide, and protein–DNA interactions that uses bacteriophages to connect proteins with the genetic information that encodes them. Phage Display was originally invented by George P...
, an alternative method for detecting protein–protein and protein–DNA interactions
- Protein array, a chip-based method for detecting protein–protein interactions
- Synthetic genetic arraySynthetic genetic arraySynthetic Genetic Array analysis is a high-throughput technique for exploring synthetic lethal and synthetic sick genetic interactions . SGA allows for the systematic construction of double mutants using a combination of recombinant genetic techniques, mating and selection steps...
analysis, a yeast based method for studying gene interactions

