MRNA surveillance
Encyclopedia
mRNA surveillance mechanisms are pathways utilized by organisms to ensure fidelity and quality of messenger RNA
(mRNA) molecules. There are a number of surveillance mechanisms present within cells. These mechanisms function at various steps of the mRNA biogenesis pathway to detect and degrade transcripts that have not properly been processed.
of messenger RNA
transcripts into proteins is a vital part of the central dogma of molecular biology
. mRNA molecules are, however, prone to a host of fidelity errors which can cause errors in translation of mRNA into quality proteins. RNA surveillance mechanisms are methods cells use to assure the quality and fidelity of the mRNA molecules. This is generally achieved through marking aberrant mRNA molecule for degradation
by various endogenous nucleases.
mRNA surveillance has been documented in bacteria
and yeast
. In eukaryotes, these mechanisms are known to function in both the nucleus
and cytoplasm
. Fidelity checks of mRNA molecules in the nucleus results in the degradation of improperly processed transcripts before export into the cytoplasm. Transcripts are subject to further surveillance once in the cytoplasm. Cytoplasmic surveillance mechanisms assess mRNA transcripts for the absence of or presence of premature stop codons .
Three surveillance mechanisms are currently known to function within cells
: the Nonsense Mediated mRNA decay pathway (NMD); the Nonstop Mediated mRNA decay pathways (NSD); and the No-go Mediated mRNA decay pathway (NGD).
mutations in DNA; somatic
mutations in DNA; errors in transcription
; or errors in post transcriptional mRNA processing. Failure to recognize and decay these mRNA transcripts can result in the production of truncated proteins which may be harmful to the organism. By causing decay of C-terminally truncated polypeptides, the NMD mechanism can protect cells against deleterious dominant-negative, and gain of function effects. PTCs have been implicated in approximately 30% of all inherited
diseases; as such, the NMD pathway plays a vital role in assuring overall survival and fitness of an organism
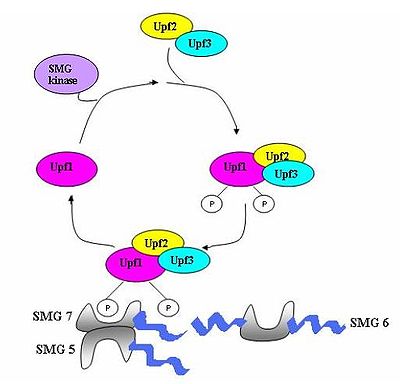
A surveillance complex consisting of various proteins (eRF1, eRF3, Upf1, Upf2 and Upf3) is assembled and scans the mRNA for premature stop codons. The assembly of this complex is triggered by premature translation termination. If a premature stop codon is detected then the mRNA transcript is signalled for degradation – the coupling of detection with degradation occurs.
Seven smg genes (smg1-7) and three UPF genes (Upf1-3) have been identified in Saccharomyces cerevisiae
and Caenorhabditis elegans
as essential trans-acting factors contributing to NMD activity. All of these genes are conserved
in Drosophila melanogaster
and further mammals where they also play critical roles in NMD. Throughout eukaryotes these are three compoments which are conserved in the process of NMD. These are the Upf1/SMG-2, Upf2/SMG-3 and Upf/SMG-4 complexes. Upf1/SMG-2 is a phosphoroprotein in mutlicellular organisms and is thought to contribute to NMD via its phosphorylation activity. However, the exact interactions of the proteins and their roles in NMD are currently disputed
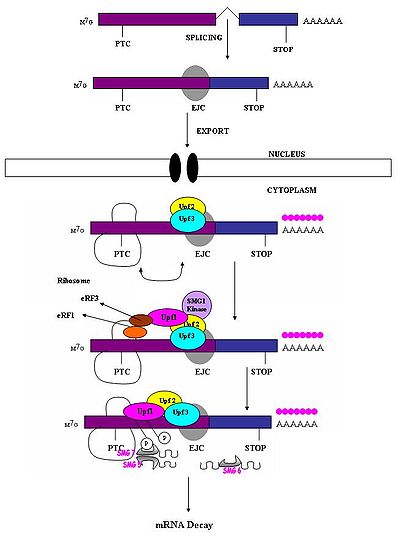 A premature stop codon must be recognized as different from a normal stop codon so that only the former triggers a NMD response. It has been observed that the ability of a nonsense codon to cause mRNA degradation depends on its relative location to the downstream sequence element and associated proteins . Studies have demonstrated that nucleotides more than 50-54 nucleotides upstream of the last exon-exon junction can target mRNA for decay. Those downstream from this region are unable to do so. Thus, nonsense codons lie more than 50-54 nucleotides upstream from the last exon
A premature stop codon must be recognized as different from a normal stop codon so that only the former triggers a NMD response. It has been observed that the ability of a nonsense codon to cause mRNA degradation depends on its relative location to the downstream sequence element and associated proteins . Studies have demonstrated that nucleotides more than 50-54 nucleotides upstream of the last exon-exon junction can target mRNA for decay. Those downstream from this region are unable to do so. Thus, nonsense codons lie more than 50-54 nucleotides upstream from the last exon
boundary whereas natural stop codons are located within terminal exons. Exon junction complex
es (EJCs) mark the exon-exon boundaries. EJCs are multiprotein complexes that assemble during splicing at a position about 20-24 nucleotides upstream from the splice junction. It is this EJC that provides position information needed to discriminate premature stop codons from natural stop codons. Recognition of PTCs appears to be dependent on the definitions of the exon-exon junctions. This suggests involvement of the spliceosome in mammalian NMD. Research has investigated the possibility of spliceosome involvement in mammalian NMD and has determined this is a likely possibility. Furthermore, it has been observed that NMD mechanisms are not activated by nonsense transcripts that are generated from genes that naturally do not contain introns (i.e. Histone H4, Hsp70, melanocortin-4-receptor).
When the ribosome
reaches a PTC the translation factors eRF1 and eRF3 interact with retained EJC complexes though a multiprotein bridge . The interactions of UPF1
with the terminating complex and with UPF2
/UPF3 of the retained EJCs are critical. It is these interactions which target the mRNA for rapid decay
by endogenous nucleases
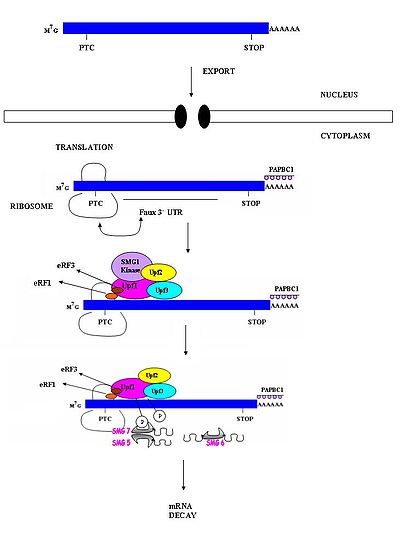
have shown that PTC recognition involving invertebrate
organisms does not involve exon-exon boundaries. These studies suggest that invertebrate NMD occurs independently of splicing. As a result, EJCs which are responsible for marking exon-exon boundaries are not required in invertebrate NMD. Several models have been proposed to explain how PTCs are distinguished from normal stop codons in invertebrates. One of these suggests that there may be a downstream sequence element which functions similar to the exon junctions in mammals. A second model proposes that a widely present feature in mRNA, such as a 3’ poly-A tail, might provide the positional information required for recognition. Another model, dubbed the “faux 3’UTR model”, suggests that premature translation termination may be distinguished from normal termination because of intrinsic features that may allow it to recognize its presence in an inappropriate environment. These mechanisms, however, have yet to be conclusively demonstrated.
This has been demonstrated in β-globulin. β-globulin mRNAs containing a nonsense mutation early in the first exon of the gene are more stable than NMD sensitive mRNA molecules. The exact mechanism of detection avoidance is currently not known. It has been suggested that the poly-A binding protein (PABP) appears to play a role in this stability. It has been demonstrated in other studies that the presence of this protein near AUG-proximal PTCs appears to promote the stability of these otherwise NMD sensitive mRNAs. It has been observed that this protective effect is not limited only to the β-globulin promoter. This suggests that this NMD avoidance mechanism may be prevalent in other tissue types for a variety of genes. The current model of NMD may need to be revisited upon further studies.
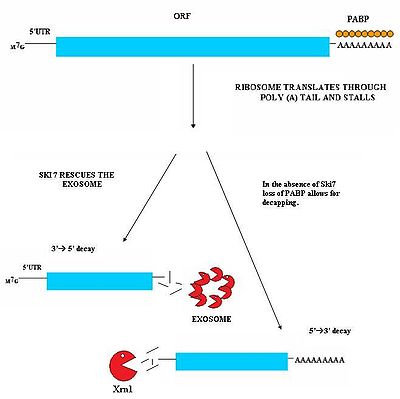 Nonstop Mediated decay (NSD) is involved in the detection and decay of mRNA transcripts which lack a stop codon. These mRNA transcripts can arise from many different mechanisms such as premature 3’ adenylation or cryptic polyadenylation signals within the coding region of a gene. This lack of a stop codon results a significant issue for cells. Ribosomes translating the mRNA eventually translate into the 3’poly-A tail region of transcripts and stalls. As a result it cannot eject the mRNA. Ribosomes thus may become sequestered associated with the nonstop mRNA and would not be available to translate other mRNA molecules into proteins. Nonstop mediated decay mediates this problem by both freeing the stalled ribosomes and marking the nonstop mRNA for degradation in the cell by nucleases. Nonstop mediated decay consists of two distinct pathways which likely act in concert to decay nonstop mRNA.
Nonstop Mediated decay (NSD) is involved in the detection and decay of mRNA transcripts which lack a stop codon. These mRNA transcripts can arise from many different mechanisms such as premature 3’ adenylation or cryptic polyadenylation signals within the coding region of a gene. This lack of a stop codon results a significant issue for cells. Ribosomes translating the mRNA eventually translate into the 3’poly-A tail region of transcripts and stalls. As a result it cannot eject the mRNA. Ribosomes thus may become sequestered associated with the nonstop mRNA and would not be available to translate other mRNA molecules into proteins. Nonstop mediated decay mediates this problem by both freeing the stalled ribosomes and marking the nonstop mRNA for degradation in the cell by nucleases. Nonstop mediated decay consists of two distinct pathways which likely act in concert to decay nonstop mRNA.
. The Ski7-exosome complex rapidly deadenylates the mRNA molecule which allows the exosome to decay the transcript in a 3’ to 5’ fashion.
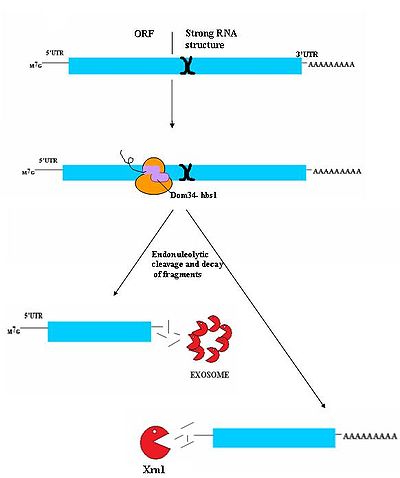 No-Go decay (NGD) is the most recently discovered surveillance mechanism. As such, it is not currently well understood. While authentic targets of NGD are poorly understood, they appear to consist largely of mRNA transcripts on which ribosomes have stalled during translation. This stall can be caused by a variety of factors including strong secondary structure
No-Go decay (NGD) is the most recently discovered surveillance mechanism. As such, it is not currently well understood. While authentic targets of NGD are poorly understood, they appear to consist largely of mRNA transcripts on which ribosomes have stalled during translation. This stall can be caused by a variety of factors including strong secondary structure
s, which may physically block the translational machinery from moving down the transcript . Dom34/Hbs1 likely binds near the A site of stalled ribosomes and may facilitate recycling of complexes . In some cases, the transcript is also cleaved in an endonucleolytic fashion near the stall site; however the identity of the responsible endonuclease remains contentious. The fragmented
mRNA molecules are then fully degraded by the exosome in a 3’ to 5’ fashion and by Xrn1 in a 5’ to 3’ fashion.
It is not currently known how this process releases the mRNA from the ribosomes, however, Hbs1 is closely related to the Ski7 protein which plays a clear role in ribosome release in Ski7 mediated NSD. It is postulated that Hbs1 may play a similar role in NGD .
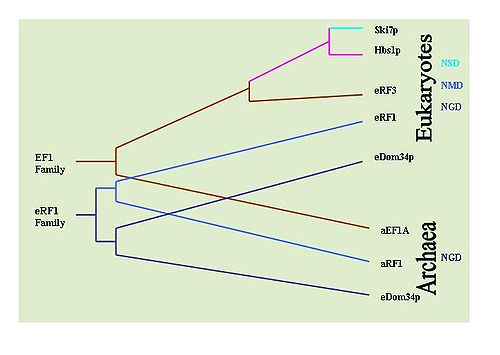
searches have been performed to determine the prevalence of the proteins in various types of organisms. It has been determined that NGD Hbs1 and NSD eRF3 are found only in eukaryotes. However, the NGD Dom34 is universal in eukaryotes and archaea
. This suggests that NGD appears to have been the first evolved mRNA surveillance mechanism. The NSD Ski7 protein appears to be restricted strictly to yeast species which suggest that NSD is the most recently evolved surveillance mechanism. This by default leaves NMD as the second evolved surveillance mechanism.
 External links=
External links=
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
(mRNA) molecules. There are a number of surveillance mechanisms present within cells. These mechanisms function at various steps of the mRNA biogenesis pathway to detect and degrade transcripts that have not properly been processed.
Overview
The translationTranslation
Translation is the communication of the meaning of a source-language text by means of an equivalent target-language text. Whereas interpreting undoubtedly antedates writing, translation began only after the appearance of written literature; there exist partial translations of the Sumerian Epic of...
of messenger RNA
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
transcripts into proteins is a vital part of the central dogma of molecular biology
Central dogma of molecular biology
The central dogma of molecular biology was first articulated by Francis Crick in 1958 and re-stated in a Nature paper published in 1970:In other words, the process of producing proteins is irreversible: a protein cannot be used to create DNA....
. mRNA molecules are, however, prone to a host of fidelity errors which can cause errors in translation of mRNA into quality proteins. RNA surveillance mechanisms are methods cells use to assure the quality and fidelity of the mRNA molecules. This is generally achieved through marking aberrant mRNA molecule for degradation
Chemical decomposition
Chemical decomposition, analysis or breakdown is the separation of a chemical compound into elements or simpler compounds. It is sometimes defined as the exact opposite of a chemical synthesis. Chemical decomposition is often an undesired chemical reaction...
by various endogenous nucleases.
mRNA surveillance has been documented in bacteria
Bacteria
Bacteria are a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a wide range of shapes, ranging from spheres to rods and spirals...
and yeast
Yeast
Yeasts are eukaryotic micro-organisms classified in the kingdom Fungi, with 1,500 species currently described estimated to be only 1% of all fungal species. Most reproduce asexually by mitosis, and many do so by an asymmetric division process called budding...
. In eukaryotes, these mechanisms are known to function in both the nucleus
Cell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
and cytoplasm
Cytoplasm
The cytoplasm is a small gel-like substance residing between the cell membrane holding all the cell's internal sub-structures , except for the nucleus. All the contents of the cells of prokaryote organisms are contained within the cytoplasm...
. Fidelity checks of mRNA molecules in the nucleus results in the degradation of improperly processed transcripts before export into the cytoplasm. Transcripts are subject to further surveillance once in the cytoplasm. Cytoplasmic surveillance mechanisms assess mRNA transcripts for the absence of or presence of premature stop codons .
Three surveillance mechanisms are currently known to function within cells
Cell (biology)
The cell is the basic structural and functional unit of all known living organisms. It is the smallest unit of life that is classified as a living thing, and is often called the building block of life. The Alberts text discusses how the "cellular building blocks" move to shape developing embryos....
: the Nonsense Mediated mRNA decay pathway (NMD); the Nonstop Mediated mRNA decay pathways (NSD); and the No-go Mediated mRNA decay pathway (NGD).
Nonsense Mediated mRNA Decay

Overview
Nonsense Mediated decay is involved in detection and decay of mRNA transcripts which contain premature termination codons (PTCs). PTCs can arise in cells through various mechanisms: germlineGermline
In biology and genetics, the germline of a mature or developing individual is the line of germ cells that have genetic material that may be passed to a child.For example, gametes such as the sperm or the egg, are part of the germline...
mutations in DNA; somatic
Somatic
The term somatic means 'of the body',, relating to the body. In medicine, somatic illness is bodily, not mental, illness. The term is often used in biology to refer to the cells of the body in contrast to the germ line cells which usually give rise to the gametes...
mutations in DNA; errors in transcription
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
; or errors in post transcriptional mRNA processing. Failure to recognize and decay these mRNA transcripts can result in the production of truncated proteins which may be harmful to the organism. By causing decay of C-terminally truncated polypeptides, the NMD mechanism can protect cells against deleterious dominant-negative, and gain of function effects. PTCs have been implicated in approximately 30% of all inherited
Heredity
Heredity is the passing of traits to offspring . This is the process by which an offspring cell or organism acquires or becomes predisposed to the characteristics of its parent cell or organism. Through heredity, variations exhibited by individuals can accumulate and cause some species to evolve...
diseases; as such, the NMD pathway plays a vital role in assuring overall survival and fitness of an organism
A surveillance complex consisting of various proteins (eRF1, eRF3, Upf1, Upf2 and Upf3) is assembled and scans the mRNA for premature stop codons. The assembly of this complex is triggered by premature translation termination. If a premature stop codon is detected then the mRNA transcript is signalled for degradation – the coupling of detection with degradation occurs.
Seven smg genes (smg1-7) and three UPF genes (Upf1-3) have been identified in Saccharomyces cerevisiae
Saccharomyces cerevisiae
Saccharomyces cerevisiae is a species of yeast. It is perhaps the most useful yeast, having been instrumental to baking and brewing since ancient times. It is believed that it was originally isolated from the skin of grapes...
and Caenorhabditis elegans
Caenorhabditis elegans
Caenorhabditis elegans is a free-living, transparent nematode , about 1 mm in length, which lives in temperate soil environments. Research into the molecular and developmental biology of C. elegans was begun in 1974 by Sydney Brenner and it has since been used extensively as a model...
as essential trans-acting factors contributing to NMD activity. All of these genes are conserved
Conserved sequence
In biology, conserved sequences are similar or identical sequences that occur within nucleic acid sequences , protein sequences, protein structures or polymeric carbohydrates across species or within different molecules produced by the same organism...
in Drosophila melanogaster
Drosophila melanogaster
Drosophila melanogaster is a species of Diptera, or the order of flies, in the family Drosophilidae. The species is known generally as the common fruit fly or vinegar fly. Starting from Charles W...
and further mammals where they also play critical roles in NMD. Throughout eukaryotes these are three compoments which are conserved in the process of NMD. These are the Upf1/SMG-2, Upf2/SMG-3 and Upf/SMG-4 complexes. Upf1/SMG-2 is a phosphoroprotein in mutlicellular organisms and is thought to contribute to NMD via its phosphorylation activity. However, the exact interactions of the proteins and their roles in NMD are currently disputed
Mechanism in mammals

Exon
An exon is a nucleic acid sequence that is represented in the mature form of an RNA molecule either after portions of a precursor RNA have been removed by cis-splicing or when two or more precursor RNA molecules have been ligated by trans-splicing. The mature RNA molecule can be a messenger RNA...
boundary whereas natural stop codons are located within terminal exons. Exon junction complex
Exon junction complex
The exon junction complex has major influences on translation, surveillance and localization of the spliced mRNA. It is first deposited onto mRNA during splicing and is then transported into the cytoplasm. There it plays a major role in post-transcriptional regulation of mRNA. It is believed that...
es (EJCs) mark the exon-exon boundaries. EJCs are multiprotein complexes that assemble during splicing at a position about 20-24 nucleotides upstream from the splice junction. It is this EJC that provides position information needed to discriminate premature stop codons from natural stop codons. Recognition of PTCs appears to be dependent on the definitions of the exon-exon junctions. This suggests involvement of the spliceosome in mammalian NMD. Research has investigated the possibility of spliceosome involvement in mammalian NMD and has determined this is a likely possibility. Furthermore, it has been observed that NMD mechanisms are not activated by nonsense transcripts that are generated from genes that naturally do not contain introns (i.e. Histone H4, Hsp70, melanocortin-4-receptor).
When the ribosome
Ribosome
A ribosome is a component of cells that assembles the twenty specific amino acid molecules to form the particular protein molecule determined by the nucleotide sequence of an RNA molecule....
reaches a PTC the translation factors eRF1 and eRF3 interact with retained EJC complexes though a multiprotein bridge . The interactions of UPF1
UPF1
Regulator of nonsense transcripts 1 is a protein that in humans is encoded by the UPF1 gene.-Interactions:UPF1 has been shown to interact with UPF2, SMG1, DCP2, DCP1A, UPF3A and UPF3B.-Further reading:...
with the terminating complex and with UPF2
UPF2
Regulator of nonsense transcripts 2 is a protein that in humans is encoded by the UPF2 gene.-Interactions:UPF2 has been shown to interact with UPF1, UPF3A and UPF3B.-Further reading:...
/UPF3 of the retained EJCs are critical. It is these interactions which target the mRNA for rapid decay
Decomposition
Decomposition is the process by which organic material is broken down into simpler forms of matter. The process is essential for recycling the finite matter that occupies physical space in the biome. Bodies of living organisms begin to decompose shortly after death...
by endogenous nucleases

Mechanism in invertebrates
Studies involving organisms such as S.cerevisiae, D.melanogaster and C. elegansCaenorhabditis elegans
Caenorhabditis elegans is a free-living, transparent nematode , about 1 mm in length, which lives in temperate soil environments. Research into the molecular and developmental biology of C. elegans was begun in 1974 by Sydney Brenner and it has since been used extensively as a model...
have shown that PTC recognition involving invertebrate
Invertebrate
An invertebrate is an animal without a backbone. The group includes 97% of all animal species – all animals except those in the chordate subphylum Vertebrata .Invertebrates form a paraphyletic group...
organisms does not involve exon-exon boundaries. These studies suggest that invertebrate NMD occurs independently of splicing. As a result, EJCs which are responsible for marking exon-exon boundaries are not required in invertebrate NMD. Several models have been proposed to explain how PTCs are distinguished from normal stop codons in invertebrates. One of these suggests that there may be a downstream sequence element which functions similar to the exon junctions in mammals. A second model proposes that a widely present feature in mRNA, such as a 3’ poly-A tail, might provide the positional information required for recognition. Another model, dubbed the “faux 3’UTR model”, suggests that premature translation termination may be distinguished from normal termination because of intrinsic features that may allow it to recognize its presence in an inappropriate environment. These mechanisms, however, have yet to be conclusively demonstrated.
NMD Avoidance
mRNAs with nonsense mutations are generally thought to be targeted for decay via the NMD pathways. The presence of this premature stop codon about 50-54 nts 5’ to the exon junction appears to be the trigger for rapid decay; however, it has been observed that some mRNA molecules with a premature stop codon are able to avoid detection and decay. In general, these mRNA molecules possess the stop codon very early in the reading frame (i.e. the PTC is AUG-proximal). This appears to be a contradiction to the current accepted model of NMD as this position is significantly 5’ of the exon-exon junction.This has been demonstrated in β-globulin. β-globulin mRNAs containing a nonsense mutation early in the first exon of the gene are more stable than NMD sensitive mRNA molecules. The exact mechanism of detection avoidance is currently not known. It has been suggested that the poly-A binding protein (PABP) appears to play a role in this stability. It has been demonstrated in other studies that the presence of this protein near AUG-proximal PTCs appears to promote the stability of these otherwise NMD sensitive mRNAs. It has been observed that this protective effect is not limited only to the β-globulin promoter. This suggests that this NMD avoidance mechanism may be prevalent in other tissue types for a variety of genes. The current model of NMD may need to be revisited upon further studies.
Overview

Ski7 Pathway
This pathway is active when Ski7 protein is available in the cell. The Ski7 protein is thought to bind to the empty A site of the ribosome. This binding allows the ribosome to eject the stuck nonstop mRNA molecule – this even frees the ribosome and allows it to translate other transcripts. The Ski7 is now associated with the nonstop mRNA and it is this association which targets the nonstop mRNA for recognition by the exosomeExosome
Exosome can refer to:* Exosome complex - a macromolecular complex involved in RNA degradation* Exosome - a vesicle secreted by mammalian cells...
. The Ski7-exosome complex rapidly deadenylates the mRNA molecule which allows the exosome to decay the transcript in a 3’ to 5’ fashion.
Non Ski7 Pathway
A second type of NMD has been observed in yeast. In this mechanism, the absence of Ski7 results in the loss of poly-A tail binding PABP proteins by the action of the translation ribosome. The removal of these PABP proteins then results in the loss of the protective 5’m7G cap. The loss of the cap results in rapid degradation of the transcript by an endogenous 5’-3’ exonuclease such as XrnI.No-Go Decay

Secondary structure
In biochemistry and structural biology, secondary structure is the general three-dimensional form of local segments of biopolymers such as proteins and nucleic acids...
s, which may physically block the translational machinery from moving down the transcript . Dom34/Hbs1 likely binds near the A site of stalled ribosomes and may facilitate recycling of complexes . In some cases, the transcript is also cleaved in an endonucleolytic fashion near the stall site; however the identity of the responsible endonuclease remains contentious. The fragmented
Fragmentation
-In biology:* Fragmentation , a form of asexual reproduction* Fragmentation * Habitat fragmentation* Population fragmentation-Music:* Fragmented , the debut album from the Filipino independent band Up Dharma Down-Other:...
mRNA molecules are then fully degraded by the exosome in a 3’ to 5’ fashion and by Xrn1 in a 5’ to 3’ fashion.
It is not currently known how this process releases the mRNA from the ribosomes, however, Hbs1 is closely related to the Ski7 protein which plays a clear role in ribosome release in Ski7 mediated NSD. It is postulated that Hbs1 may play a similar role in NGD .
Evolution of mRNA Surveillance Mechanisms
It is possible to determine the evolutionary history of these mechanisms by observing the conservation of key proteins implicated in each mechanism. For example: Dom34/Hbs1 are associated with NDG[31]; Ski7 is associated with NSD; and the eRF proteins are associated with NMD. To this end, extensive BLASTBLAST
In bioinformatics, Basic Local Alignment Search Tool, or BLAST, is an algorithm for comparing primary biological sequence information, such as the amino-acid sequences of different proteins or the nucleotides of DNA sequences...
searches have been performed to determine the prevalence of the proteins in various types of organisms. It has been determined that NGD Hbs1 and NSD eRF3 are found only in eukaryotes. However, the NGD Dom34 is universal in eukaryotes and archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
. This suggests that NGD appears to have been the first evolved mRNA surveillance mechanism. The NSD Ski7 protein appears to be restricted strictly to yeast species which suggest that NSD is the most recently evolved surveillance mechanism. This by default leaves NMD as the second evolved surveillance mechanism.


