Chip-Sequencing
Encyclopedia
ChIP-Sequencing, also known as ChIP-Seq, is used to analyze protein
interactions with DNA
. ChIP-Seq combines chromatin immunoprecipitation
(ChIP) with massively parallel DNA sequencing
to identify the cistrome
of DNA-associated proteins. It can be used to precisely map global binding sites for any protein of interest. Previously, ChIP-on-chip
was the most common technique utilized to study these protein–DNA relations.
 Chip-Seq is used primarily to determine how transcription factors and other chromatin-associated proteins influence phenotype
Chip-Seq is used primarily to determine how transcription factors and other chromatin-associated proteins influence phenotype
-affecting mechanisms. Determining how proteins interact with DNA to regulate gene expression
is essential for fully understanding many biological processes and disease states. This epigenetic information is complementary to genotype
and expression analysis. ChIP-Seq technology is currently seen primarily as an alternative to ChIP-chip which requires a hybridization array. This necessarily introduces some bias, as an array is restricted to a fixed number of probes. Sequencing, by contrast, is thought to have less bias, although the sequencing bias of different sequencing technologies is not yet fully understood.
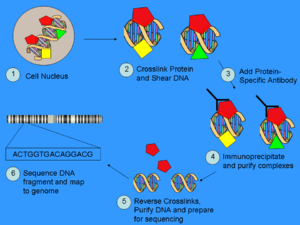
Specific DNA sites in direct physical interaction with transcription factors and other proteins can be isolated by chromatin immunoprecipitation
. ChIP produces a library of target DNA sites bound to a target in vivo
. Massively parallel sequence analyses are used in conjunction with whole-genome sequence databases to analyze the interaction pattern of any protein with DNA, or the pattern of any epigenetic chromatin
modifications. This can be applied to the set of ChIP-able proteins and modifications, such as transcription factors, polymerase
s and transcriptional machinery, structural proteins, protein modifications, and DNA modifications. As an alternative to the dependence on specific antibodies, different methods have been developed to find the superset of all nucleosome-depleted or nucleosome-disrupted active regulatory regions in the genome, like DNase-Seq
and FAIRE-Seq
.
is a powerful method to selectively enrich for DNA sequences bound by a particular protein in living cells
. However, the widespread use of this method has been limited by the lack of a sufficiently robust method to identify all of the enriched DNA sequences. The ChIP process enriches specific crosslinked DNA-protein complexes using an antibody
against a protein of interest. For a good description of the ChIP wet lab protocol see the ChIP-on-chip
Wikipedia page. Oligonucleotide
adapters are then added to the small stretches of DNA that were bound to the protein of interest to enable massively parallel sequencing.
s for lower resolution.
There are many new sequencing methods used in this sequencing step. Some technologies that analyze the sequences can use cluster amplification of adapter-ligated ChIP DNA fragments on a solid flow cell substrate to create clusters of approximately 1000 clonal copies each. The resulting high density array of template clusters on the flow cell surface is sequenced by a Genome analyzing program. Each template cluster undergoes sequencing-by-synthesis in parallel using novel fluorescently labelled reversible terminator nucleotides. Templates are sequenced base-by-base during each read. Then, the data collection and analysis software aligns sample sequences to a known genomic sequence to identify the ChIP-DNA fragments.
The sequencing depth is directly correlated with cost. If abundant binders in large genomes have to be mapped with high sensitivity, costs are high as an enormously high number of sequence tags will be required. This is in contrast to ChIP-chip in which the costs are not correlated with sensitivity.
Unlike microarray
-based ChIP methods, the precision of the ChIP-Seq assay is not limited by the spacing of predetermined probes. By integrating a large number of short reads, highly precise binding site localization is obtained. Compared to ChIP-chip, ChIP-Seq data can be used to locate the binding site within few tens of base pairs of the actual protein binding site. Tag densities at the binding sites are a good indicator of protein–DNA binding affinity, which makes it easier to quantify and compare binding affinities of a protein to different DNA sites.
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
interactions with DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
. ChIP-Seq combines chromatin immunoprecipitation
Chromatin immunoprecipitation
Chromatin Immunoprecipitation is a type of immunoprecipitation experimental technique used to investigate the interaction between proteins and DNA in the cell. It aims to determine whether specific proteins are associated with specific genomic regions, such as transcription factors on promoters or...
(ChIP) with massively parallel DNA sequencing
DNA sequencing
DNA sequencing includes several methods and technologies that are used for determining the order of the nucleotide bases—adenine, guanine, cytosine, and thymine—in a molecule of DNA....
to identify the cistrome
Cistrome
CistromeThis term http://cistrome.pbwiki.com was coined by investigators at the Dana-Farber Cancer Institute and Harvard Medical School to define the set of cis-acting targets of a trans-acting factor on a genome scale...
of DNA-associated proteins. It can be used to precisely map global binding sites for any protein of interest. Previously, ChIP-on-chip
ChIP-on-chip
ChIP-on-chip is a technique that combines chromatin immunoprecipitation with microarray technology . Like regular ChIP, ChIP-on-chip is used to investigate interactions between proteins and DNA in vivo...
was the most common technique utilized to study these protein–DNA relations.
Uses of ChIP-Seq

Phenotype
A phenotype is an organism's observable characteristics or traits: such as its morphology, development, biochemical or physiological properties, behavior, and products of behavior...
-affecting mechanisms. Determining how proteins interact with DNA to regulate gene expression
Gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA , transfer RNA or small nuclear RNA genes, the product is a functional RNA...
is essential for fully understanding many biological processes and disease states. This epigenetic information is complementary to genotype
Genotype
The genotype is the genetic makeup of a cell, an organism, or an individual usually with reference to a specific character under consideration...
and expression analysis. ChIP-Seq technology is currently seen primarily as an alternative to ChIP-chip which requires a hybridization array. This necessarily introduces some bias, as an array is restricted to a fixed number of probes. Sequencing, by contrast, is thought to have less bias, although the sequencing bias of different sequencing technologies is not yet fully understood.
Specific DNA sites in direct physical interaction with transcription factors and other proteins can be isolated by chromatin immunoprecipitation
Chromatin immunoprecipitation
Chromatin Immunoprecipitation is a type of immunoprecipitation experimental technique used to investigate the interaction between proteins and DNA in the cell. It aims to determine whether specific proteins are associated with specific genomic regions, such as transcription factors on promoters or...
. ChIP produces a library of target DNA sites bound to a target in vivo
In vivo
In vivo is experimentation using a whole, living organism as opposed to a partial or dead organism, or an in vitro controlled environment. Animal testing and clinical trials are two forms of in vivo research...
. Massively parallel sequence analyses are used in conjunction with whole-genome sequence databases to analyze the interaction pattern of any protein with DNA, or the pattern of any epigenetic chromatin
Chromatin
Chromatin is the combination of DNA and proteins that make up the contents of the nucleus of a cell. The primary functions of chromatin are; to package DNA into a smaller volume to fit in the cell, to strengthen the DNA to allow mitosis and meiosis and prevent DNA damage, and to control gene...
modifications. This can be applied to the set of ChIP-able proteins and modifications, such as transcription factors, polymerase
Polymerase
A polymerase is an enzyme whose central function is associated with polymers of nucleic acids such as RNA and DNA.The primary function of a polymerase is the polymerization of new DNA or RNA against an existing DNA or RNA template in the processes of replication and transcription...
s and transcriptional machinery, structural proteins, protein modifications, and DNA modifications. As an alternative to the dependence on specific antibodies, different methods have been developed to find the superset of all nucleosome-depleted or nucleosome-disrupted active regulatory regions in the genome, like DNase-Seq
DNase-Seq
DNase-Seq is a method in molecular biology used to identify the location of regulatory regions, based on the genome-wide sequencing of regions super sensitivity to cleavage by DNase I. FAIRE-Seq is a successor of DNase-Seq for the genome-wide identification of accessible DNA regions in the genome....
and FAIRE-Seq
FAIRE-Seq
FAIRE-Seq is a method in molecular biology used for determining the sequences of those DNA regions in the genome associated with regulatory activity. In contrast to DNase-seq, the FAIRE-Seq protocol doesn't require the permeabilization of cells or isolation of nuclei, and can analyse any cell types...
.
Part 1: ChIP
ChIPChip
- Food :* Potato chips , a snack food made from potatoes, also known as crisps in the UK and some other English-speaking countries...
is a powerful method to selectively enrich for DNA sequences bound by a particular protein in living cells
Cell (biology)
The cell is the basic structural and functional unit of all known living organisms. It is the smallest unit of life that is classified as a living thing, and is often called the building block of life. The Alberts text discusses how the "cellular building blocks" move to shape developing embryos....
. However, the widespread use of this method has been limited by the lack of a sufficiently robust method to identify all of the enriched DNA sequences. The ChIP process enriches specific crosslinked DNA-protein complexes using an antibody
Antibody
An antibody, also known as an immunoglobulin, is a large Y-shaped protein used by the immune system to identify and neutralize foreign objects such as bacteria and viruses. The antibody recognizes a unique part of the foreign target, termed an antigen...
against a protein of interest. For a good description of the ChIP wet lab protocol see the ChIP-on-chip
ChIP-on-chip
ChIP-on-chip is a technique that combines chromatin immunoprecipitation with microarray technology . Like regular ChIP, ChIP-on-chip is used to investigate interactions between proteins and DNA in vivo...
Wikipedia page. Oligonucleotide
Oligonucleotide
An oligonucleotide is a short nucleic acid polymer, typically with fifty or fewer bases. Although they can be formed by bond cleavage of longer segments, they are now more commonly synthesized, in a sequence-specific manner, from individual nucleoside phosphoramidites...
adapters are then added to the small stretches of DNA that were bound to the protein of interest to enable massively parallel sequencing.
Part 2: Sequencing
After size selection, all the resulting ChIP-DNA fragments are sequenced simultaneously using a genome sequencer. A single sequencing run can scan for genome-wide associations with high resolution, meaning that features can be located precisely on the chromosomes. ChIP-chip, by contrast, requires large sets of tiling arrayTiling array
Tiling Arrays are a subtype of microarray chips. Like traditional microarrays, they function by hybridizing labeled DNA or RNA target molecules to probes fixed onto a solid surface. Tiling arrays differ from traditional microarrays in the nature of the probes...
s for lower resolution.
There are many new sequencing methods used in this sequencing step. Some technologies that analyze the sequences can use cluster amplification of adapter-ligated ChIP DNA fragments on a solid flow cell substrate to create clusters of approximately 1000 clonal copies each. The resulting high density array of template clusters on the flow cell surface is sequenced by a Genome analyzing program. Each template cluster undergoes sequencing-by-synthesis in parallel using novel fluorescently labelled reversible terminator nucleotides. Templates are sequenced base-by-base during each read. Then, the data collection and analysis software aligns sample sequences to a known genomic sequence to identify the ChIP-DNA fragments.
Sensitivity
Sensitivity of this technology depends on the depth of the sequencing run (i.e. the number of mapped sequence tags), the size of the genome and the distribution of the target factor.The sequencing depth is directly correlated with cost. If abundant binders in large genomes have to be mapped with high sensitivity, costs are high as an enormously high number of sequence tags will be required. This is in contrast to ChIP-chip in which the costs are not correlated with sensitivity.
Unlike microarray
Microarray
A microarray is a multiplex lab-on-a-chip. It is a 2D array on a solid substrate that assays large amounts of biological material using high-throughput screening methods.Types of microarrays include:...
-based ChIP methods, the precision of the ChIP-Seq assay is not limited by the spacing of predetermined probes. By integrating a large number of short reads, highly precise binding site localization is obtained. Compared to ChIP-chip, ChIP-Seq data can be used to locate the binding site within few tens of base pairs of the actual protein binding site. Tag densities at the binding sites are a good indicator of protein–DNA binding affinity, which makes it easier to quantify and compare binding affinities of a protein to different DNA sites.
Current Research
- STAT1 DNA association: Recently, ChIP-Seq was used to study STAT1 targets in HeLA S3 cells. The performance of ChIP-Seq was then compared to the alternative protein–DNA interaction methods of ChIP-PCR and ChIP-chip.
- Nucleosome Architecture of Promoters: Using ChIP-Seq, it was determined that Yeast genes seem to have a minimal nucleosome-free promoter region of 150bp in which RNA polymerase can initiate transcription.
Conclusion
In summary, ChIP-seq offers an alternative to ChIP-chip. STAT1 experimental ChIP-seq data have a high degree of similarity to results obtained by ChIP-chip for the same type of experiment, with >64% of peaks in shared genomic regions. Because the data are sequence reads, ChIP-seq offers a rapid analysis pipeline (as long as a high-quality genome sequence is available for read mapping, and the genome doesn't have repetitive content that confuses the mapping process) as well as the potential to detect mutations in binding-site sequences, which may directly support any observed changes in protein binding and gene regulation.Similar methods
- Sono-SeqSono-SeqSono-Seq is a method in molecular biology used for determining the sequences of those DNA regions in the genome near regions of open chromatin of expressed genes. It is also known as "Input" in the Chip-Seq protocol, since it follows the same steps except it doesn't require immunoprecipitation....
, identical to Chip-Seq but skipping the immunoprecipitation step. - CLIP-SeqCLIP-SeqCLIP-Seq, also called HITS-CLIP, is a method in molecular biology, used for finding which RNA species interact with a particular RNA-binding protein...
, for finding interactions with RNA rather than DNA. - PAR-CLIPPAR-CLIPPAR-CLIP is a novel method, used for identifying the binding sites of cellular RNA-binding proteins and microRNA-containing ribonucleoprotein complexes...
, for identifying the binding sites of cellular RNA-binding proteins (RBPs) and microRNA-containing ribonucleoprotein complexes (miRNPs). - RIP-ChipRIP-ChipRIP-Chip is immunoprecipitation of an RNA-binding protein coupled to reverse transcription and a microarray. It has been used to find interactions between RNA and protein ....
, same goal and first steps, but using microarray instead of sequencing - SELEXSystematic Evolution of Ligands by Exponential EnrichmentSELEX , also referred to as in vitro selection or in vitro evolution, is a combinatorial chemistry technique in molecular biology for producing oligonucleotides of either single-stranded DNA or RNA that specifically bind to a target ligand or ligands....
, a method for finding a consensus binding sequence - Competition-ChIPCompetition-ChIPCompetition-ChIP is variant of the Chip-Sequencing protocol, used to measure relative binding dynamics of a transcription factor on DNA. Since TF occupancy measures are thought to be a poor predictor of TF function at a given locus, Competition-ChIP is much more strongly linked to function than...
, to measure relative replacement dynamics on DNA. - ChiRP-SeqChiRP-SeqChIRP-Seq is a high-throughput sequencing method to discover RNA-bound DNA and proteins. The RNA sequences of interest are hybridized to oligonucleotide tiles using biotin/streptavidin. The chromatin fraction that is bound to the beads is then determined using high-throughput sequencing....
to measure RNA-bound DNA and proteins.

