
ChIP-on-chip
Encyclopedia
ChIP-on-chip is a technique that combines chromatin immunoprecipitation
Chromatin immunoprecipitation
Chromatin Immunoprecipitation is a type of immunoprecipitation experimental technique used to investigate the interaction between proteins and DNA in the cell. It aims to determine whether specific proteins are associated with specific genomic regions, such as transcription factors on promoters or...
("ChIP") with microarray technology
DNA microarray
A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome...
("chip"). Like regular ChIP, ChIP-on-chip is used to investigate interactions between protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s and DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
in vivo
In vivo
In vivo is experimentation using a whole, living organism as opposed to a partial or dead organism, or an in vitro controlled environment. Animal testing and clinical trials are two forms of in vivo research...
. Specifically, it allows the identification of the cistrome
Cistrome
CistromeThis term http://cistrome.pbwiki.com was coined by investigators at the Dana-Farber Cancer Institute and Harvard Medical School to define the set of cis-acting targets of a trans-acting factor on a genome scale...
, sum of binding site
Binding site
In biochemistry, a binding site is a region on a protein, DNA, or RNA to which specific other molecules and ions—in this context collectively called ligands—form a chemical bond...
s, for DNA-binding proteins on a genome-wide basis. Whole-genome analysis can be performed to determine the locations of binding sites for almost any protein of interest. As the name of the technique suggests, such proteins are generally those operating in the context of chromatin
Chromatin
Chromatin is the combination of DNA and proteins that make up the contents of the nucleus of a cell. The primary functions of chromatin are; to package DNA into a smaller volume to fit in the cell, to strengthen the DNA to allow mitosis and meiosis and prevent DNA damage, and to control gene...
. The most prominent representatives of this class are transcription factor
Transcription factor
In molecular biology and genetics, a transcription factor is a protein that binds to specific DNA sequences, thereby controlling the flow of genetic information from DNA to mRNA...
s, replication
DNA replication
DNA replication is a biological process that occurs in all living organisms and copies their DNA; it is the basis for biological inheritance. The process starts with one double-stranded DNA molecule and produces two identical copies of the molecule...
-related proteins, like ORC, histone
Histone
In biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
s, their variants, and histone modifications.
The goal of ChIP-on-chip is to localize protein binding sites that may help identify functional elements in the genome
Genome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
. For example, in the case of a transcription factor as a protein of interest, one can determine its transcription factor binding sites throughout the genome. Other proteins allow the identification of promoter regions, enhancers
Enhancer (genetics)
In genetics, an enhancer is a short region of DNA that can be bound with proteins to enhance transcription levels of genes in a gene cluster...
, repressors and silencing elements
Silencer (DNA)
In genetics a silencer is a DNA sequence capable of binding transcription regulation factors termed repressors. Upon binding, RNA polymerase is prevented from initiating transcription thus decreasing or fully suppressing RNA synthesis....
, insulators, boundary elements, and sequences that control DNA replication. If histones are subject of interest, it is believed that the distribution of modifications and their localizations may offer new insights into the mechanisms of regulation.
One of the long-term goals ChIP-on-chip was designed for is to establish a catalogue of (selected) organisms that lists all protein-DNA interaction
Protein-DNA interaction
Protein–DNA interactions are when a protein binds a molecule of DNA, often to regulate the biological function of DNA, usually the expression of a gene...
s under various physiological conditions. This knowledge would ultimately help in the understanding of the machinery behind gene regulation, cell proliferation, and disease progression. Hence, ChIP-on-chip offers not only huge potential to complement our knowledge about the orchestration of the genome on the nucleotide level, but also on higher levels of information and regulation as it is propagated by research on epigenetics
Epigenetics
In biology, and specifically genetics, epigenetics is the study of heritable changes in gene expression or cellular phenotype caused by mechanisms other than changes in the underlying DNA sequence – hence the name epi- -genetics...
.
Technological platforms
The technical platforms to conduct ChIP-on-chip experiments are DNA microarrayDNA microarray
A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome...
s, or "chips". They can be classified and distinguished according to various characteristics:
- Probe type: DNA arrays can comprise either mechanically spotted cDNAs or PCR-products, mechanically spotted oligonucleotideOligonucleotideAn oligonucleotide is a short nucleic acid polymer, typically with fifty or fewer bases. Although they can be formed by bond cleavage of longer segments, they are now more commonly synthesized, in a sequence-specific manner, from individual nucleoside phosphoramidites...
s, or oligonucleotides that are synthesized in situIn situIn situ is a Latin phrase which translated literally as 'In position'. It is used in many different contexts.-Aerospace:In the aerospace industry, equipment on board aircraft must be tested in situ, or in place, to confirm everything functions properly as a system. Individually, each piece may...
. The early versions of microarrays were designed to detect RNARNARibonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
s from expressed genomic regions (open reading frameOpen reading frameIn molecular genetics, an open reading frame is a DNA sequence that does not contain a stop codon in a given reading frame.Normally, inserts which interrupt the reading frame of a subsequent region after the start codon cause frameshift mutation of the sequence and dislocate the sequences for stop...
s). Although such arrays are perfectly suited to study gene expression profilesExpression profilingIn the field of molecular biology, gene expression profiling is the measurement of the activity of thousands of genes at once, to create a global picture of cellular function. These profiles can, for example, distinguish between cells that are actively dividing, or show how the cells react to a...
, they have limited importance in ChIP experiments since most "interesting" proteins with respect to this technique bind in intergenic regionIntergenic regionAn Intergenic region is a stretch of DNA sequences located between clusters of genes that contain few or no genes. Occasionally some intergenic DNA acts to control genes nearby, but most of it has no currently known function...
s. Nowadays, even custom-made arrays can be designed and fine-tuned to match the requirements of an experiment. Also, any sequence of nucleotides can be synthesized to cover genic as well as intergenic regions. - Probe size: Early version of cDNA arrays had a probe length of about 200bp. Latest array versions use oligos as short as 70- (Microarrays, Inc.) to 25-mers (AffymetrixAffymetrixAffymetrix is a company that manufactures DNA microarrays; it is based in Santa Clara, California, United States. The company was founded by Dr. Stephen Fodor in 1992. It began as a unit in Affymax N.V...
). (Feb 2007) - Probe composition: There are tiled and non-tiled DNA arrays. Non-tiled arrays use probes selected according to non-spatial criteria, i.e., the DNA sequences used as probes have no fixed distances in the genome. Tiled arrays, however, select a genomic region (or even a whole genome) and divide it into equal chunks. Such a region is called tiled path. The average distance between each pair of neighboring chunks (measured from the center of each chunk) gives the resolution of the tiled path. A path can be overlapping, end-to-end or spaced.
- Array size: The first microarrays used for ChIP-on-Chip contained about 13,000 spotted DNA segments representing all ORFs and intergenic regions from the yeast genome. Nowadays, Affymetrix offers whole-genome tiled yeast arrays with a resolution of 5bp (all in all 3.2 million probes). Tiled arrays for the human genome become more and more powerful, too. Just to name example, Affymetrix offers a set of seven arrays with about 90 million probes, spanning the complete non-repetitive part of the human genome with about 35bp spacing. (Feb 2007)
Besides the actual microarray, other hard- and software equipment is necessary to run ChIP-on-chip experiments. It is generally the case that one company’s microarrays can not be analyzed by another company’s processing hardware. Hence, buying an array requires also buying the associated workflow equipment. The most important elements are, among others, hybridization ovens, chip scanners, and software packages for subsequent numerical analysis of the raw data.
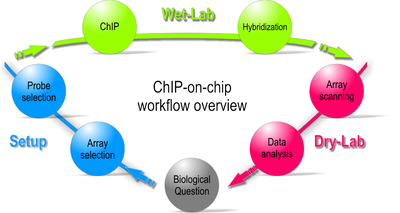
Workflow of a ChIP-on-chip experiment
Starting with a biological question, a ChIP-on-chip experiment can be divided into three major steps: The first is to set up and design the experiment by selecting the appropriate array and probe type. Second, the actual experiment is performed in the wet-lab. Last, during the dry-lab portion of the cycle, gathered data are analyzed to either answer the initial question or lead to new questions so that the cycle can start again.Wet-lab portion of the workflow

- In the first step, the protein of interest (POI) is cross-linkCross-linkCross-links are bonds that link one polymer chain to another. They can be covalent bonds or ionic bonds. "Polymer chains" can refer to synthetic polymers or natural polymers . When the term "cross-linking" is used in the synthetic polymer science field, it usually refers to the use of...
ed with the DNA site it binds to in an in vivoIn vivoIn vivo is experimentation using a whole, living organism as opposed to a partial or dead organism, or an in vitro controlled environment. Animal testing and clinical trials are two forms of in vivo research...
environment. Usually this is done by a gentle formaldehydeFormaldehydeFormaldehyde is an organic compound with the formula CH2O. It is the simplest aldehyde, hence its systematic name methanal.Formaldehyde is a colorless gas with a characteristic pungent odor. It is an important precursor to many other chemical compounds, especially for polymers...
fixation that is reversible with heat. - Then, the cells are lysed and the DNA is sheared by sonicationSonicationthumb|right|A sonicator at the [[Weizmann Institute of Science]] during sonicationSonication is the act of applying sound energy to agitate particles in a sample, for various purposes. In the laboratory, it is usually applied using an ultrasonic bath or an ultrasonic probe, colloquially known as...
or using micrococcal nucleaseMicrococcal nucleaseMicrococcal Nuclease is an endo-exonuclease that preferentially digests single-stranded nucleic acids.The rate of cleavage is 30 times greater at the 5' side of A or T than at G or C and results in the production of mononucleotides and oligonucleotides with terminal 3'-phosphates...
. This results in double-stranded chunks of DNA fragments, normally 1 kb or less in length. Those that were cross-linkCross-linkCross-links are bonds that link one polymer chain to another. They can be covalent bonds or ionic bonds. "Polymer chains" can refer to synthetic polymers or natural polymers . When the term "cross-linking" is used in the synthetic polymer science field, it usually refers to the use of...
ed to the POI form a POI-DNA complex. - In the next step, only these complexes are filtered out of the set of DNA fragments, using an antibodyAntibodyAn antibody, also known as an immunoglobulin, is a large Y-shaped protein used by the immune system to identify and neutralize foreign objects such as bacteria and viruses. The antibody recognizes a unique part of the foreign target, termed an antigen...
specific to the POI. The antibodies may be attached to a solid surface, may have a magnetic bead, or some other physical property that allow distributing cross-linked complexes and unbound fragments. This procedure is essentially an immunoprecipitationImmunoprecipitationImmunoprecipitation is the technique of precipitating a protein antigen out of solution using an antibody that specifically binds to that particular protein. This process can be used to isolate and concentrate a particular protein from a sample containing many thousands of different proteins...
(IP). There are two alternative ways to implement this filtering step:- immunoprecipitation of the tagged protein with an antibody against the tag (ex. FLAGFLAG-tagFLAG-tag, or FLAG octapeptide, is a polypeptide protein tag that can be added to a protein using recombinant DNA technology. It can be used for affinity chromatography, then used to separate recombinant, overexpressed protein from wild-type protein expressed by the host organism...
, HAHemagglutininInfluenza hemagglutinin or haemagglutinin is a type of hemagglutinin found on the surface of the influenza viruses. It is an antigenic glycoprotein. It is responsible for binding the virus to the cell that is being infected...
, c-myc) - affinity purification that does not require antibodies, such as the Tandem Affinity PurificationTandem Affinity PurificationTandem affinity purification is a technique for studying protein-protein interactions. It involves creating a fusion protein with a designed piece, the TAP tag, on the end...
(TAP)
- immunoprecipitation of the tagged protein with an antibody against the tag (ex. FLAG
- The cross-linking of POI-DNA complexes is reversed (usually by heating) and the DNA strands are purified. For the rest of the workflow, the POI is no longer necessary.
- After an amplification and denaturationDenaturation (biochemistry)Denaturation is a process in which proteins or nucleic acids lose their tertiary structure and secondary structure by application of some external stress or compound, such as a strong acid or base, a concentrated inorganic salt, an organic solvent , or heat...
step, the single-stranded DNA fragments are labeled with a fluorescentFluorescenceFluorescence is the emission of light by a substance that has absorbed light or other electromagnetic radiation of a different wavelength. It is a form of luminescence. In most cases, emitted light has a longer wavelength, and therefore lower energy, than the absorbed radiation...
tag such as Cy5 or Alexa 647. - Finally, the fragments are poured over the surface of the DNA microarray, which is spotted with short, single-stranded sequences that cover the genomic portion of interest. Whenever a labeled fragment "finds" a complementary fragment on the array, they will hybridize and form again a double-stranded DNA fragment.
- Quantitative and qualitative DNA and protein analysis with very small volumes of valuable samples are essential steps during the whole wet-lab workflow. Specialized NanoPhotometer offer the possibility of UV-visible spectrophotometry with sample volumes of 0.3 µl. Reduction of the optical pathlength, compared to standard measurements, effects an automatic dilution of the samples. Therefore, samples can be measured undiluted and if desired, retrieved after the measurement for further processing.
Dry-lab portion of the workflow
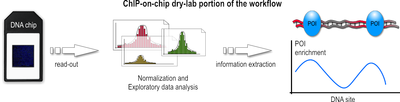
- After a sufficiently large time frame to allow hybridization, the array is illuminated with fluorescence light. Those probes on the array that are hybridized to one of the labeled fragments emit a light signal that is captured by a camera. This image contains all raw data for the remaining part of the workflow.
- This raw data, encoded as false-color imageFalse-colorA false-color image is an image that depicts a subject in colors that differ from those a full-color photograph would show.-True- and false-color:...
, needs to be converted to numerical values before the actual analysis can be done. The analysis and information extraction of the raw data often remains the most challenging part for ChIP-on-chip experiments. Problems arise throughout this portion of the workflow, ranging from the initial chip read-out, to suitable methods to subtract background noise, and finally to appropriate algorithmAlgorithmIn mathematics and computer science, an algorithm is an effective method expressed as a finite list of well-defined instructions for calculating a function. Algorithms are used for calculation, data processing, and automated reasoning...
s that normalizeNormalization (statistics)In one usage in statistics, normalization is the process of isolating statistical error in repeated measured data. A normalization is sometimes based on a property...
the data and make it available for subsequent statistical analysisStatisticsStatistics is the study of the collection, organization, analysis, and interpretation of data. It deals with all aspects of this, including the planning of data collection in terms of the design of surveys and experiments....
, which then hopefully lead to a better understanding of the biological question sought to answer. Furthermore, due to the different array platforms and missing standardization between them, data storage and exchange is a huge problem, too. Generally speaking, the data analysis can be divided into three major steps:
- During the first step, the captured fluorescence signals from the array are normalized, using control signals derived from the same or a second chip. Such control signals tell which probes on the array were hybridized correctly and which bound nonspecifically.
- In the second step, numerical and statistical tests are applied to control data and IP fraction data to identify POI-enriched regions along the genome. The following three methods are used widely: Median percentile rank, Single-array error, and Sliding-window. These methods generally differ in a way how low-intensity signals are handled, how much background noise is accepted, and which trait for the data is emphasized during the computation. In the recent past, the sliding-window approach seems to be favored and is often described as most powerful.
- In the third step, these regions are analyzed further. If, for example, the POI was a transcription factor, such regions would represent its binding sites. Subsequent analysis then may want to infer nucleotide motifs and other patterns to allow functional annotation of the genome.
Strengths and Weaknesses
Using tiled arraysDNA microarray
A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome...
, ChIP
Chip
- Food :* Potato chips , a snack food made from potatoes, also known as crisps in the UK and some other English-speaking countries...
-on-chip allows for high resolution of genome-wide maps. These maps can determine the binding sites of many DNA-binding proteins like transcription factors and also chromatin modifications.
Although ChIP-on-chip can be a powerful technique in the area of genomics, it is very expensive. Most published studies using ChIP-on-chip repeat their experiments at least three times to ensure biologically meaningful maps. The cost of the DNA microarrays is often a limiting factor to whether a laboratory should proceed with a ChIP-on-chip experiment. Another limitation is the size of DNA fragments that can be achieved. Most ChIP-on-chip protocols utilize sonication as a method of breaking up DNA into small pieces. However, sonication is limited to a minimal fragment size of 200 bp. For higher resolution maps, this limitation should be overcome to achieve smaller fragments, preferably to single nucleosome
Nucleosome
Nucleosomes are the basic unit of DNA packaging in eukaryotes, consisting of a segment of DNA wound around a histone protein core. This structure is often compared to thread wrapped around a spool....
resolution. As mentioned previously, the statistical analysis of the huge amount of data generated from arrays is a challenge and normalization procedures should aim to minimize artifacts and determine what is really biologically significant. So far, application to mammalian genomes has been a major limitation, for example, due to a significant percentage of the genome that is occupied by repeats. However, as ChIP-on-chip technology advances, high resolution whole mammalian genome maps should become achievable.
Antibodies
Antibody
An antibody, also known as an immunoglobulin, is a large Y-shaped protein used by the immune system to identify and neutralize foreign objects such as bacteria and viruses. The antibody recognizes a unique part of the foreign target, termed an antigen...
used for ChIP
Chip
- Food :* Potato chips , a snack food made from potatoes, also known as crisps in the UK and some other English-speaking countries...
-on-chip can be an important limiting factor. ChIP
Chip
- Food :* Potato chips , a snack food made from potatoes, also known as crisps in the UK and some other English-speaking countries...
-on-chip requires highly specific antibodies that must recognize its epitope
Epitope
An epitope, also known as antigenic determinant, is the part of an antigen that is recognized by the immune system, specifically by antibodies, B cells, or T cells. The part of an antibody that recognizes the epitope is called a paratope...
in free solution and also under fixed conditions. If it is demonstrated to successfully immunoprecipitate cross-linked chromatin
Chromatin
Chromatin is the combination of DNA and proteins that make up the contents of the nucleus of a cell. The primary functions of chromatin are; to package DNA into a smaller volume to fit in the cell, to strengthen the DNA to allow mitosis and meiosis and prevent DNA damage, and to control gene...
, it is termed "ChIP-grade". Companies that provide ChIP-grade antibodies include Abcam
Abcam plc
Abcam plc is a UK biotech company based in the Cambridge Science Park in Cambridge, England, with offices in Boston MA, San Francisco, Hong Kong and Tokyo.The premise of the company was incubated from a laboratory in the University of Cambridge, UK in 1998...
, Cell Signaling Technology
Cell Signaling Technology
Cell Signaling Technology, Inc. established in Beverly, Massachusetts in 1999, is a privately-held company which produces antibodies and related reagents used to study cell signaling pathways. CST's current corporate headquarters are located in Danvers, Massachusetts.-History:Cell Signaling...
, Santa Cruz, and Upstate. To overcome the problem of specificity, the protein of interest can be fused to a tag like FLAG
FLAG-tag
FLAG-tag, or FLAG octapeptide, is a polypeptide protein tag that can be added to a protein using recombinant DNA technology. It can be used for affinity chromatography, then used to separate recombinant, overexpressed protein from wild-type protein expressed by the host organism...
or HA
Hemagglutinin
Influenza hemagglutinin or haemagglutinin is a type of hemagglutinin found on the surface of the influenza viruses. It is an antigenic glycoprotein. It is responsible for binding the virus to the cell that is being infected...
that are recognized by antibodies. An alternative to ChIP-on-chip that does not require antibodies is DamID
DamID
DamID is a molecular biology protocol used to map the binding sites of DNA- and chromatin-binding proteins in eukaryotes. DamID identifies binding sites by expressing the proposed DNA-binding protein as a fusion protein with DNA methyltransferase...
.
Also available are antibodies against a specific histone modification like H3
Histone H3
Histone H3 is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a string' structure...
tri methyl K4. As mentioned before, the combination of these antibodies and ChIP-on-chip has become extremely powerful in determining whole genome analysis of histone modification patterns and will contribute tremendously to our understanding of the histone code
Histone code
The histone code is a hypothesis that the transcription of genetic information encoded in DNA is in part regulated by chemical modifications to histone proteins, primarily on their unstructured ends. Together with similar modifications such as DNA methylation it is part of the epigenetic code...
and epigenetics.
A study demonstrating the non-specific nature of DNA binding proteins has been published in PLoS Biology. This indicates that alternate confirmation of functional relevancy is a necessary step in any ChIP-chip experiment.
History
A first ChIP-on-chip experiment was performed in 1999 to analyze the distribution of cohesin along buddingBudding
Budding is a form of asexual reproduction in which a new organism grows on another one. The new organism remains attached as it grows, separating from the parent organism only when it is mature. Since the reproduction is asexual, the newly created organism is a clone and is genetically identical...
yeast
Yeast
Yeasts are eukaryotic micro-organisms classified in the kingdom Fungi, with 1,500 species currently described estimated to be only 1% of all fungal species. Most reproduce asexually by mitosis, and many do so by an asymmetric division process called budding...
chromosome III. Although the genome was not completely represented, the protocol in this study remains equivalent as those used in later studies. The ChIP-on-chip technique using all of the ORFs of the genome (that nevertheless remains incomplete, missing intergenic regions) was then applied successfully in three papers published in 2000 and 2001. The authors identified binding sites for individual transcription factors in the budding
Budding
Budding is a form of asexual reproduction in which a new organism grows on another one. The new organism remains attached as it grows, separating from the parent organism only when it is mature. Since the reproduction is asexual, the newly created organism is a clone and is genetically identical...
yeast
Yeast
Yeasts are eukaryotic micro-organisms classified in the kingdom Fungi, with 1,500 species currently described estimated to be only 1% of all fungal species. Most reproduce asexually by mitosis, and many do so by an asymmetric division process called budding...
Saccharomyces cerevisiae
Saccharomyces cerevisiae
Saccharomyces cerevisiae is a species of yeast. It is perhaps the most useful yeast, having been instrumental to baking and brewing since ancient times. It is believed that it was originally isolated from the skin of grapes...
. In 2002, Richard Young’s group determined the genome-wide positions of 106 transcription factors using a c-Myc tagging system in yeast. Other applications for ChIP-on-chip include DNA replication
DNA replication
DNA replication is a biological process that occurs in all living organisms and copies their DNA; it is the basis for biological inheritance. The process starts with one double-stranded DNA molecule and produces two identical copies of the molecule...
, recombination
Genetic recombination
Genetic recombination is a process by which a molecule of nucleic acid is broken and then joined to a different one. Recombination can occur between similar molecules of DNA, as in homologous recombination, or dissimilar molecules, as in non-homologous end joining. Recombination is a common method...
, and chromatin structure. Since then, ChIP-on-chip has become a powerful tool in determining genome-wide maps of histone modifications and many more transcription factors. ChIP-on-chip in mammalian systems has been difficult due to the large and repetitive genomes. Thus, many studies in mammalian cells have focused on select promoter regions that are predicted to bind transcription factors and have not analyzed the entire genome. However, whole mammalian genome arrays have recently become commercially available from companies like Nimblegen. In the future, as ChIP-on-chip arrays become more and more advanced, high resolution whole genome maps of DNA-binding proteins and chromatin components for mammals will be analyzed in more detail.
Analysis and Software
http://www.ciml.univ-mrs.fr/software/cocas/index.html CoCAS: a free Analysis software for Agilent ChIP-on-Chip experimentshttp://www.bioconductor.org/packages/2.4/bioc/html/rMAT.html rMAT: R implementation from MAT program to normalize and analyze tiling arrays and ChIP-chip data.
Software Reference
http://bioinformatics.oxfordjournals.org/cgi/content/abstract/btp075 Touati Benoukraf , Pierre Cauchy , Romain Fenouil , Adrien Jeanniard , Frederic Koch , Sébastien Jaeger , Denis Thieffry , Jean Imbert , Jean-Christophe Andrau , Salvatore Spicuglia , and Pierre Ferrier , CoCAS: a ChIP-on-chip analysis suite, Bioinformatics Advance Access published on April 1, 2009, DOI 10.1093/bioinformatics/btp075, Bioinformatics 25: 954-955.http://www.pnas.org/content/103/33/12457.abstract W. Evan Johnson, Wei Li, Clifford A. Meyer, Raphael Gottardo, Jason S. Carroll, Myles Brown, and X. Shirley Liu. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci U S A. 2006 Aug 15;103(33):12457-62. Epub 2006 Aug 8.
Alternatives
Chip-SequencingChip-Sequencing
ChIP-Sequencing, also known as ChIP-Seq, is used to analyze protein interactions with DNA. ChIP-Seq combines chromatin immunoprecipitation with massively parallel DNA sequencing to identify the cistrome of DNA-associated proteins. It can be used to precisely map global binding sites for any...
is a recently developed technology that still uses chromatin immunoprecipitation to crosslink the proteins of interest to the DNA but then instead of using a micro-array, it uses the more accurate, higher throughput method of sequencing to localize interaction points.
DamID
DamID
DamID is a molecular biology protocol used to map the binding sites of DNA- and chromatin-binding proteins in eukaryotes. DamID identifies binding sites by expressing the proposed DNA-binding protein as a fusion protein with DNA methyltransferase...
is an alternative method that does not require antibodies.
External links
- http://www.genome.gov/10005107 ENCODE project
- http://www.chiponchip.org
- List of ChIP-chip software Wikiomics@OpenWetWare
- Affymetrix Chip-on-Chip Forum
- http://chipanalysis.genomecenter.ucdavis.edu/cgi-bin/tamalpais.cgi Online analysis of NimbleGen ChIP-chip experiments

