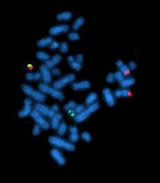
Fluorescent in situ hybridization
Encyclopedia
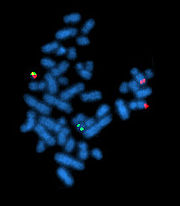
Cytogenetics
Cytogenetics is a branch of genetics that is concerned with the study of the structure and function of the cell, especially the chromosomes. It includes routine analysis of G-Banded chromosomes, other cytogenetic banding techniques, as well as molecular cytogenetics such as fluorescent in situ...
technique developed by biomedical researchers in the early 1980s that is used to detect and localize the presence or absence of specific DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
sequences
DNA sequence
The sequence or primary structure of a nucleic acid is the composition of atoms that make up the nucleic acid and the chemical bonds that bond those atoms. Because nucleic acids, such as DNA and RNA, are unbranched polymers, this specification is equivalent to specifying the sequence of...
on chromosome
Chromosome
A chromosome is an organized structure of DNA and protein found in cells. It is a single piece of coiled DNA containing many genes, regulatory elements and other nucleotide sequences. Chromosomes also contain DNA-bound proteins, which serve to package the DNA and control its functions.Chromosomes...
s. FISH uses fluorescent probes
Hybridization probe
In molecular biology, a hybridization probe is a fragment of DNA or RNA of variable length , which is used in DNA or RNA samples to detect the presence of nucleotide sequences that are complementary to the sequence in the probe...
that bind to only those parts of the chromosome with which they show a high degree of sequence complementarity. Fluorescence microscopy can be used to find out where the fluorescent probe bound to the chromosomes. FISH is often used for finding specific features in DNA for use in genetic counselling, medicine, and species identification. FISH can also be used to detect and localize specific mRNAs within tissue samples. In this context, it can help define the spatial-temporal patterns of gene expression
Gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA , transfer RNA or small nuclear RNA genes, the product is a functional RNA...
within cells and tissues.
Probes
Size exclusion chromatography
Size-exclusion chromatography is a chromatographic method in which molecules in solution are separated by their size, and in some cases molecular weight . It is usually applied to large molecules or macromolecular complexes such as proteins and industrial polymers...
, and using that information to determine where the large fragments overlapped one another.) To preserve the fragments with their individual DNA sequences, the fragments were added into a system of continually replicating bacteria populations. Clonal populations of bacteria, each population maintaining a single artificial chromosome, are stored in various laboratories around the world. The artificial chromosomes (BAC
Bacterial artificial chromosome
A bacterial artificial chromosome is a DNA construct, based on a functional fertility plasmid , used for transforming and cloning in bacteria, usually E. coli. F-plasmids play a crucial role because they contain partition genes that promote the even distribution of plasmids after bacterial cell...
) can be grown, extracted, and labeled, in any lab. These fragments are on the order of 100 thousand base-pairs, and are the basis for most FISH probes.
Preparation and Hybridization Process
.jpg)
Fluorescent tag
In molecular biology and biotechnology, a fluorescent tag is a part of a molecule that researchers have attached chemically to aid in detection of the molecule to which it has been attached. The tag is some kind of fluorescent molecule...
directly with fluorophore
Fluorophore
A fluorophore, in analogy to a chromophore, is a component of a molecule which causes a molecule to be fluorescent. It is a functional group in a molecule which will absorb energy of a specific wavelength and re-emit energy at a different wavelength...
s, with targets for antibodies or with biotin
Biotin
Biotin, also known as Vitamin H or Coenzyme R, is a water-soluble B-complex vitamin discovered by Bateman in 1916. It is composed of a ureido ring fused with a tetrahydrothiophene ring. A valeric acid substituent is attached to one of the carbon atoms of the tetrahydrothiophene ring...
. Tagging can be done in various ways, such as nick translation
Nick translation
Nick translation was developed in 1977 by Rigby and Paul Berg. It is a tagging technique in molecular biology in which DNA Polymerase I is used to replace some of the nucleotides of a DNA sequence with their labeled analogues, creating a tagged DNA sequence which can be used as a probe in...
, or PCR using tagged nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
s.
Then, an interphase
Interphase
Interphase is the phase of the cell cycle in which the cell spends the majority of its time and performs the majority of its purposes including preparation for cell division. In preparation for cell division, it increases its size and makes a copy of its DNA...
or metaphase
Metaphase
Metaphase, from the ancient Greek μετά and φάσις , is a stage of mitosis in the eukaryotic cell cycle in which condensed & highly coiled chromosomes, carrying genetic information, align in the middle of the cell before being separated into each of the two daughter cells...
chromosome preparation is produced. The chromosomes are firmly attached to a substrate
Substrate (biochemistry)
In biochemistry, a substrate is a molecule upon which an enzyme acts. Enzymes catalyze chemical reactions involving the substrate. In the case of a single substrate, the substrate binds with the enzyme active site, and an enzyme-substrate complex is formed. The substrate is transformed into one or...
, usually glass. Repetitive DNA sequences must be blocked by adding short fragments of DNA to the sample. The probe is then applied to the chromosome DNA and incubated for approximately 12 hours while hybridizing. Several wash steps remove all unhybridized or partially-hybridized probes. The results are then visualized and quantified using a microscope that is capable of exciting the dye and recording images.
If the fluorescent signal is weak, amplification of the signal may be necessary in order to exceed the detection threshold of the microscope
Microscope
A microscope is an instrument used to see objects that are too small for the naked eye. The science of investigating small objects using such an instrument is called microscopy...
. Fluorescent signal strength depends on many factors such as probe labeling efficiency, the type of probe, and the type of dye. The dye incorporation rate can be quantified photometric with specialized nano-volume photometer (starting with 0.3 µl probe volume). Fluorescently-tagged antibodies or streptavidin
Streptavidin
Streptavidin is a 60000 dalton protein purified from the bacterium Streptomyces avidinii. Streptavidin homo-tetramers have an extraordinarily high affinity for biotin . With a dissociation constant on the order of ≈10-14 mol/L, the binding of biotin to streptavidin is one of the strongest...
are bound to the dye molecule. These secondary components are selected so that they have a strong signal.
FISH experiments designed to detect or localize gene expression
Gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA , transfer RNA or small nuclear RNA genes, the product is a functional RNA...
within cells and tissues rely on the use of a reporter gene
Reporter gene
In molecular biology, a reporter gene is a gene that researchers attach to a regulatory sequence of another gene of interest in cell culture, animals or plants. Certain genes are chosen as reporters because the characteristics they confer on organisms expressing them are easily identified and...
, such as one expressing green fluorescent protein
Green fluorescent protein
The green fluorescent protein is a protein composed of 238 amino acid residues that exhibits bright green fluorescence when exposed to blue light. Although many other marine organisms have similar green fluorescent proteins, GFP traditionally refers to the protein first isolated from the...
, to provide the fluorescence signal.
Variations on probes and analysis
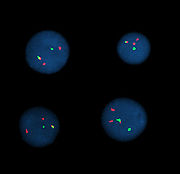
Probe size is important because longer probes hybridize less specifically than shorter probes. The overlap defines the resolution of detectable features. For example, if the goal of an experiment is to detect the breakpoint of a translocation
Translocation
Translocation may refer to:* Chromosomal translocation, in genetics* Translocation in plants, transport of food or pesticides through phloem or xylem* Protein translocation or protein targeting, a process in protein biosynthesis...
, then the overlap of the probes — the degree to which one DNA sequence is contained in the adjacent probes — defines the minimum window in which the breakpoint may be detected.
The mixture of probe sequences determines the type of feature the probe can detect. Probes that hybridize along an entire chromosome are used to count the number of a certain chromosome, show translocations, or identify extra-chromosomal fragments of chromatin
Chromatin
Chromatin is the combination of DNA and proteins that make up the contents of the nucleus of a cell. The primary functions of chromatin are; to package DNA into a smaller volume to fit in the cell, to strengthen the DNA to allow mitosis and meiosis and prevent DNA damage, and to control gene...
. This is often called "whole-chromosome painting." If every possible probe is used, every chromosome, (the whole genome) would be marked fluorescently, which would not be particularly useful for determining features of individual sequences. However, a mixture of smaller probes can be created that is specific to a particular region (locus) of DNA; these mixtures are used to detect deletion mutations
Genetic deletion
In genetics, a deletion is a mutation in which a part of a chromosome or a sequence of DNA is missing. Deletion is the loss of genetic material. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome...
. When combined with a specific colour, a locus-specific probe mixture is used to detect very specific translocations. Special locus-specific probe mixtures are often used to count chromosomes, by binding to the centromeric regions of chromosomes, which are unique enough to identify each chromosome (with the exception of Chromosome 13, 14, 21, 22.)
A variety of other techniques use mixtures of differently-colored probes. A range of colors in mixtures of fluorescent dyes can be detected, so each human chromosome can be identified by a characteristic color using whole-chromosome probe mixtures and a variety of ratios of colors. Although there are more chromosomes than easily-distinguishable fluorescent dye colors, ratios of probe mixtures can be used to create secondary colors. Similar to comparative genomic hybridization
Comparative genomic hybridization
Comparative genomic hybridization or Chromosomal Microarray Analysis is a molecular-cytogenetic method for the analysis of copy number changes in the DNA content of a given subject's DNA and often in tumor cells....
, the probe mixture for the secondary colors is created by mixing the correct ratio of two sets of differently-colored probes for the same chromosome. This technique is sometimes called M-FISH. The same physics that make a variety of colors possible for M-FISH can be used for the detection of translocations. That is, colors that are adjacent appear to overlap; a secondary color is observed. Some assays are designed so that the secondary color will be present or absent in cases of interest. An example is the detection of BCR/ABL
Philadelphia chromosome
Philadelphia chromosome or Philadelphia translocation is a specific chromosomal abnormality that is associated with chronic myelogenous leukemia . It is the result of a reciprocal translocation between chromosome 9 and 22, and is specifically designated t...
translocations, where the secondary color indicates disease. This variation is often called double-fusion FISH or D-FISH. In the opposite situation---where the absence of the secondary color is pathological---is illustrated by an assay used to investigate translocations where only one of the breakpoints is known or constant. Locus-specific probes are made for one side of the breakpoint and the other intact chromosome. In normal cells, the secondary colour is observed, but only the primary colour is observed when the translocation occurs. This technique is sometimes called "break-apart FISH".
Stellaris FISH Probes
Stellaris FISH, formerly known as Single Molecule RNA FISH, is a method of detecting and quantifying mRNA and other long RNA molecules in a thin layer of tissue sample. Targets can be reliably imaged through the application of multiple short singly labeled oligonucleotide probesHybridization probe
In molecular biology, a hybridization probe is a fragment of DNA or RNA of variable length , which is used in DNA or RNA samples to detect the presence of nucleotide sequences that are complementary to the sequence in the probe...
. The binding of up to 48 fluorescent labeled oligos to a single molecule of mRNA provides sufficient fluorescence to accurately detect and localize each target mRNA in a wide-field fluorescent microscopy image. Probes not binding to the intended sequence do not achieve sufficient localized fluorescence to be distinguished from background
Background noise
In acoustics and specifically in acoustical engineering, background noise or ambient noise is any sound other than the sound being monitored. Background noise is a form of noise pollution or interference. Background noise is an important concept in setting noise regulations...
.
Single molecule RNA FISH assays can be performed in simplex or multiplex
Multiplex (assay)
A multiplex assay is a type of laboratory procedure that simultaneously measures multiple analytes in a single assay. It is distinguished from procedures that measure one or a few analytes at a time...
, and can be used as a follow-up experiment to qPCR, or imaged simultaneously with a fluorescent antibody
Immunohistochemistry
Immunohistochemistry or IHC refers to the process of detecting antigens in cells of a tissue section by exploiting the principle of antibodies binding specifically to antigens in biological tissues. IHC takes its name from the roots "immuno," in reference to antibodies used in the procedure, and...
assay. The technology has potential applications in cancer diagnosis, neuroscience
Neuroscience
Neuroscience is the scientific study of the nervous system. Traditionally, neuroscience has been seen as a branch of biology. However, it is currently an interdisciplinary science that collaborates with other fields such as chemistry, computer science, engineering, linguistics, mathematics,...
, gene expression
Gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA , transfer RNA or small nuclear RNA genes, the product is a functional RNA...
analysis, and companion diagnostics
Personalized medicine
Personalized medicine is a medical model emphasizing in general the customization of healthcare, with all decisions and practices being tailored to individual patients in whatever ways possible...
.
Fiber FISH
In an alternative technique to interphase or metaphase preparations, fiber FISH, interphaseInterphase
Interphase is the phase of the cell cycle in which the cell spends the majority of its time and performs the majority of its purposes including preparation for cell division. In preparation for cell division, it increases its size and makes a copy of its DNA...
chromosomes are attached to a slide in such a way that they are stretched out in a straight line, rather than being tightly coiled, as in conventional FISH, or adopting a random conformation, as in interphase FISH. This is accomplished by applying mechanical shear
Shearing (physics)
Shearing in continuum mechanics refers to the occurrence of a shear strain, which is a deformation of a material substance in which parallel internal surfaces slide past one another. It is induced by a shear stress in the material...
along the length of the slide, either to cells that have been fixed to the slide and then lysed
Lysis
Lysis refers to the breaking down of a cell, often by viral, enzymic, or osmotic mechanisms that compromise its integrity. A fluid containing the contents of lysed cells is called a "lysate"....
, or to a solution of purified DNA. A technique known as chromosome combing
Chromosome combing
Chromosome combing is a technique used to produce an array of uniformly stretched DNA that is then highly suitable for nucleic acid hybridization studies such as fluorescent in situ hybridisation which benefit from the uniformity of stretching, the easy access to the hybridisation target...
is increasingly used for this purpose. The extended conformation of the chromosomes allows dramatically higher resolution - even down to a few kilobases. The preparation of fiber FISH samples, although conceptually simple, is a rather skilled art, and only specialized laboratories use the technique routinely.
Q-FISH
Q-FISHQ-FISH
Quantitative Fluorescent in situ hybridization is a cytogenetic technique based on the traditional FISH methodology. In Q-FISH, the technique uses labelled synthetic DNA mimics called peptide nucleic acid oligonucleotides to quantify target sequences in chromosomal DNA using fluorescent...
combines FISH with PNAs and computer software to quantify fluorescence intensity. This technique is used routinely in telomere
Telomere
A telomere is a region of repetitive DNA sequences at the end of a chromosome, which protects the end of the chromosome from deterioration or from fusion with neighboring chromosomes. Its name is derived from the Greek nouns telos "end" and merοs "part"...
length research.
Flow-FISH
Flow-FISHFlow-FISH
Flow-FISH is a cytogenetic technique to quantify the copy number of specific repetitive elements in genomic DNA of whole cell populations via the combination of flow cytometry with cytogenetic fluorescent in situ hybridization staining protocols...
uses flow cytometry
Flow cytometry
Flow cytometry is a technique for counting and examining microscopic particles, such as cells and chromosomes, by suspending them in a stream of fluid and passing them by an electronic detection apparatus. It allows simultaneous multiparametric analysis of the physical and/or chemical...
to perform FISH automatically using per-cell fluorescence measurements.
Medical applications
Often parents of children with a developmental delay want to know more about their child's conditions before choosing to have another child. These concerns can be addressed by analysis of the parents' and child's DNA. In cases where the child's developmental delay is not understood, the cause of it can potentially be determined using FISH and cytogeneticCytogenetics
Cytogenetics is a branch of genetics that is concerned with the study of the structure and function of the cell, especially the chromosomes. It includes routine analysis of G-Banded chromosomes, other cytogenetic banding techniques, as well as molecular cytogenetics such as fluorescent in situ...
techniques. Examples of diseases that are diagnosed using FISH include Prader-Willi syndrome
Prader-Willi syndrome
Prader–Willi syndrome is a rare genetic disorder in which seven genes on chromosome 15 are deleted or unexpressed on the paternal chromosome...
, Angelman syndrome
Angelman syndrome
Angelman syndrome is a neuro-genetic disorder characterized by intellectual and developmental delay, sleep disturbance, seizures, jerky movements , frequent laughter or smiling, and usually a happy demeanor....
, 22q13 deletion syndrome
22q13 deletion syndrome
22q13 Deletion Syndrome , also known as Phelan-McDermid Syndrome, is a genetic disorder caused by a microdeletion on chromosome 22. The deletion occurs at the terminal end of the chromosome at the location designated q13.3...
, chronic myelogenous leukemia
Chronic myelogenous leukemia
Chronic myelogenous leukemia , also known as chronic granulocytic leukemia , is a cancer of the white blood cells. It is a form of leukemia characterized by the increased and unregulated growth of predominantly myeloid cells in the bone marrow and the accumulation of these cells in the blood...
, acute lymphoblastic leukemia
Acute lymphoblastic leukemia
Acute lymphoblastic leukemia is a form of leukemia, or cancer of the white blood cells characterized by excess lymphoblasts.Malignant, immature white blood cells continuously multiply and are overproduced in the bone marrow. ALL causes damage and death by crowding out normal cells in the bone...
, Cri-du-chat, Velocardiofacial syndrome, and Down syndrome
Down syndrome
Down syndrome, or Down's syndrome, trisomy 21, is a chromosomal condition caused by the presence of all or part of an extra 21st chromosome. It is named after John Langdon Down, the British physician who described the syndrome in 1866. The condition was clinically described earlier in the 19th...
. FISH on sperm cells is indicated for men with an abnormal somatic or meiotic karyotype
Karyotype
A karyotype is the number and appearance of chromosomes in the nucleus of an eukaryotic cell. The term is also used for the complete set of chromosomes in a species, or an individual organism.p28...
as well as those with oligozoospermia, since approximately 50% of oligozoospermic men have an increased rate of sperm chromosome abnormalities. The analysis of chromosomes 21, X, and Y is enough to identify oligozoospermic individuals at risk.
In medicine, FISH can be used to form a diagnosis
Diagnosis
Diagnosis is the identification of the nature and cause of anything. Diagnosis is used in many different disciplines with variations in the use of logics, analytics, and experience to determine the cause and effect relationships...
, to evaluate prognosis
Prognosis
Prognosis is a medical term to describe the likely outcome of an illness.When applied to large statistical populations, prognostic estimates can be very accurate: for example the statement "45% of patients with severe septic shock will die within 28 days" can be made with some confidence, because...
, or to evaluate remission of a disease, such as cancer
Cancer
Cancer , known medically as a malignant neoplasm, is a large group of different diseases, all involving unregulated cell growth. In cancer, cells divide and grow uncontrollably, forming malignant tumors, and invade nearby parts of the body. The cancer may also spread to more distant parts of the...
. Treatment can then be specifically tailored. A traditional exam involving metaphase chromosome analysis is often unable to identify features that distinguish one disease from another, due to subtle chromosomal features; FISH can elucidate these differences. FISH can also be used to detect diseased cells more easily than standard Cytogenetic methods, which require dividing cells and requires labor and time-intensive manual preparation and analysis of the slides by a technologist. FISH, on the other hand, does not require living cells and can be quantified automatically, a computer counts the fluorescent dots present. However, a trained technologist is required to distinguish subtle differences in banding patterns on bent and twisted metaphase chromosomes.
Species identification
FISH is often used in clinical studies. If a patient is infected with a suspected pathogenPathogen
A pathogen gignomai "I give birth to") or infectious agent — colloquially, a germ — is a microbe or microorganism such as a virus, bacterium, prion, or fungus that causes disease in its animal or plant host...
, bacteria, from the patient's tissues or fluids, are typically grown on agar to determine the identity of the pathogen. Many bacteria, however, even well-known species, do not grow well under laboratory conditions. FISH can be used to detect directly the presence of the suspect on small samples of patient's tissue.
FISH can also be used to compare the genomes of two biological species, to deduce evolution
Evolution
Evolution is any change across successive generations in the heritable characteristics of biological populations. Evolutionary processes give rise to diversity at every level of biological organisation, including species, individual organisms and molecules such as DNA and proteins.Life on Earth...
ary relationships. A similar hybridization technique is called a zoo blot
Zoo blot
A zoo blot or garden blot is a type of Southern blot that demonstrates the similarity between specific, usually protein-coding, DNA sequences of different species...
. Bacterial FISH probes are often primers for the 16s rRNA region.
FISH is widely used in the field of microbial ecology
Microbial ecology
Microbial ecology is the ecology of microorganisms: their relationship with one another and with their environment. It concerns the three major domains of life — Eukaryota, Archaea, and Bacteria — as well as viruses....
, to identify microorganisms. Biofilms, for example, are composed of complex (often) multi-species bacterial organizations. Preparing DNA probes for one species and performing FISH with this probe allows one to visualize the distribution of this specific species within the biofilm. Preparing probes (in two different colors) for two species allows to visualize/study co-localization of these two species in the biofilm, and can be useful in determining the fine architecture of the biofilm.
Lab-on-a-chip and FISH

Currently, FISH has been performed on glass microfluidic platforms that standardize much of the protocol offering repeatable results that are accurate, cost-effective and easier to obtain in a clinical setting.
Compared to conventional FISH methods, these first implementations of on-chip FISH provide a 10-fold higher throughput and a 10-fold reduction in the cost of testing, enabling the simultaneous assessment of several chromosomal abnormalities or patients. It is increasingly essential that diagnostic tests determine the type and extent of chromosomal abnormalities for more informed diagnosis and for appropriate choice of treatment strategies. Since the on-chip FISH technique is 10-20 times more cost-effective than conventional methods, and can be fully integrated and automated, this technology will make widespread genetic testing of patients more accessible in a clinical setting.
Recently, the first demonstration of Metaphase FISH on chip has led to renewed efforts towards automating the metaphase FISH protocol. Metaphase FISH had continued to be difficult to integrate owing to the complex sample preparation protocol often spanning over 3 weeks. New reports confirm that a research group in Denmark have tested successfully a novel lab on chip device to integrate the entire sample preparation protocol for Metaphase FISH called FISHprep.
Virtual Karyotype
Virtual karyotypingVirtual Karyotype
Virtual karyotype detects genomic copy number variations at a higher resolution level than conventional karyotyping or chromosome-based comparative genomic hybridization .-Background:...
is another cost-effective, clinically available alternative to FISH panels uses thousands to millions of probes on a single array to detect copy number changes, genome-wide, at unprecedented resolution. Currently, this type of analysis will only detect gains and losses of chromosomal material and will not detect balanced rearrangements, such as translocations and inversions which are hallmark aberrations seen in many types of leukemia and lymphoma.
See also
- In situ hybridizationIn situ hybridizationIn situ hybridization is a type of hybridization that uses a labeled complementary DNA or RNA strand to localize a specific DNA or RNA sequence in a portion or section of tissue , or, if the tissue is small enough , in the entire tissue...
- Molecular cytogeneticsMolecular cytogeneticsMolecular cytogenetics involves the combination of molecular biology and cytogenetics. In general this involves the use of a series of techniques referred to as fluorescence in situ hybridization, or FISH, in which DNA probes are labeled with different colored fluorescent tags to visualize one or...
- Virtual KaryotypeVirtual KaryotypeVirtual karyotype detects genomic copy number variations at a higher resolution level than conventional karyotyping or chromosome-based comparative genomic hybridization .-Background:...
- Happy mappingHappy mappingHAPPY Mapping, by Paul H. Dear and Peter R. Cook in 1989, is a method used in molecular biology to study the linkage between two or more DNA sequences. According to the , it is "Mapping based on the analysis of approximately HAPloid DNA samples using the PolYmerase chain reaction"...
External links
- Information on fiber FISH from the Olympus CorporationOlympus Corporationis a Japan-based manufacturer of optics and reprography products. Olympus was established on 12 October 1919, initially specializing in microscope and thermometer businesses. Its global headquarters are in Shinjuku, Tokyo, Japan, while its USA operations are based in Center Valley, Pennsylvania,...
- A guide to fiber FISH from Octavian Henegariu
- Fibre FISH protocol from the Human Genome ProjectHuman Genome ProjectThe Human Genome Project is an international scientific research project with a primary goal of determining the sequence of chemical base pairs which make up DNA, and of identifying and mapping the approximately 20,000–25,000 genes of the human genome from both a physical and functional...
at the Sanger Centre - CARD-FISH, BioMineWiki
- Preparation of Complex DNA Probe Sets for 3D FISH with up to Six Different Fluorochromes
- FISH technical notes and protocols from GeneDetect.com
- Fluorescence in situ Hybridization Photos of bacteria
- Rational design of polynucleotide probe mixes to identify particular genes in defined taxa: www.dnaBaser.com/PolyPro

