
Halogen bond
Encyclopedia
Halogen bonding is the non-covalent interaction that occurs between a halogen
atom (Lewis acid) and a Lewis base. Although halogens are involved in other types of bonding (e.g. covalent), halogen bonding specifically refers to when the halogen acts as an electrophilic species.
In both cases, D (donor) is the atom, group, or molecule that donates the electron poor species (H or X). H is the hydrogen atom involved in HB, and X is the halogen atom involved in XB. A (acceptor) is the electron rich species.
A parallel relationship can easily be drawn between halogen bonding and hydrogen bonding (HB). In both types of bonding, an electron donor
/electron acceptor
relationship exists. The difference between the two is what species can act as the electron donor/electron acceptor. In hydrogen bonding, a hydrogen atom acts as the electron acceptor and forms a non-covalent interaction by accepting electron density
from an electron rich site (electron donor). In halogen bonding, a halogen atom is the electron acceptor. Electron density transfers results in a penetration of the van der Waals
volumes.
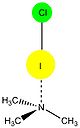 Halogens participating in halogen bonding include: iodine
Halogens participating in halogen bonding include: iodine
(I), bromine
(Br), chlorine
(Cl), and sometimes fluorine
(F). All four halogens are capable of acting as XB donors (as proven through theoretical and experimental data) and follow the general trend: F < Cl < Br < I, with iodine normally forming the strongest interactions.
Dihalogens (I2, Br2, etc.) tend to form strong halogen bonds. The strength and effectiveness of chlorine and fluorine in XB formation depend on the nature of the XB donor. If the halogen is bonded to an electronegative (electron withdrawing) moiety, it is more likely to form stronger halogen bonds.
For example, iodoperfluoroalkanes are well-designed for XB crystal engineering
. In addition, this is also why F2 can act as a strong XB donor, but fluorocarbon
s are weak XB donors because the alkyl group connected to the fluorine is not electronegative. In addition, the Lewis base (XB acceptor) tends to be electronegative as well and anions are better XB acceptors than neutral molecules.
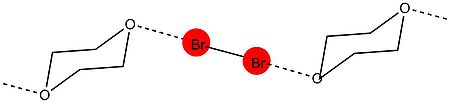
Halogen bonds are strong, specific, and directional interactions that give rise to well-defined structures. Halogen bond strengths range from 5–180 kJ/mol. The strength of XB allows it to compete with HB, which are a little bit weaker in strength. Halogen bonds tend to form at 180° angles, which was shown in Odd Hassel
’s studies with bromine and 1,4-dioxane in 1954. Another contributing factor to halogen bond strength comes from the short distance between the halogen (Lewis acid, XB donor) and Lewis base (XB acceptor). The attractive nature of halogen bonds result in the distance between the donor and acceptor to be shorter than the sum of van der Waals radii. The XB interaction becomes stronger as the distance decreases between the halogen and Lewis base.
gave the first report on the ability of halogen atoms to form well-defined adducts with electron donor species. In his experiment, he added I2 to a saturated solution of ammonium nitrate to form NH3I2. When the compound was exposed to air, it spontaneously decomposed into ammonia
and iodine which allowed Guthrie to conclude that he had formed NH3I2.
In the 1950s, Robert S. Mulliken
developed a detailed theory of electron donor-acceptor complexes, classifying them as being outer or inner complexes. Outer complexes were those in which the intermolecular interaction between the electron donor and acceptor were weak and had very little charge transfer. Inner complexes have extensive charge redistribution. Mulliken’s theory has been used to describe the mechanism by which XB formation occurs.
 Around the same time period that Mulliken developed his theory, crystallographic studies performed by Hassel began to emerge and became a turning point in the comprehension of XB formation and its characteristics.
Around the same time period that Mulliken developed his theory, crystallographic studies performed by Hassel began to emerge and became a turning point in the comprehension of XB formation and its characteristics.
The first X-ray crystallography study from Hassel’s group came in 1954. In the experiment, his group was able to show the structure of bromine 1,4-dioxanate using x-ray diffraction techniques. The experiment revealed that a short intermolecular interaction was present between the oxygen atoms of dioxane and bromine atoms. The O−Br distance in the crystal was measured at 2.71 Å, which indicates a strong interaction between the bromine and oxygen atoms. In addition, the distance is smaller than the sum of the van der Waals radii of oxygen and bromine (3.35 Å). The angle between the O−Br and Br−Br bond is about 180°. This was the first evidence of the typical characteristics found in halogen bond formation and led Hassel to conclude that halogen atoms are directly linked to electron pair donor with a bond direction that coincides with the axes of the orbitals of the lone pairs in the electron pair donor molecule.
In the 1980s continued work was carried out using analytical methods such as infrared spectroscopy
and Fourier transform spectroscopy
. These methods allowed the isolation of complexes formed between Lewis bases and halogen molecules for further studies.
is a growing research area that bridges solid-state and supramolecular chemistry. This unique field is interdisciplinary and merges traditional disciplines such as crystallography
, organic chemistry
, and inorganic chemistry
. In 1971, Schmidt first established the field with a publication on photodimerization in the solid-state. The more recent definition identifies crystal engineering as the utilization of the intermolecular interactions for crystallization and for the development of new substances with different desired physicochemical properties. Before the discovery of halogen bonding, the approach for crystal engineering involved using hydrogen bonding, coordination chemistry and inter-ion interactions for the development of liquid-crystalline and solid-crystalline materials. Furthermore, halogen bonding is employed for the organization of radical cationic salts, fabrication of molecular conductors, and creation of liquid crystal constructs. Since the discovery halogen bonding, new molecular assemblies exist. Due to the unique chemical nature of halogen bonding, this intermolecular interaction serves as an additional tool for the development of crystal engineering.
The first reported use of halogen bonding in liquid crystal formation was by H. Loc Nguyen. In an effort to form liquid crystals, alkoxystilbazoles and pentafluoroiodobenzene were used. Previous studies by Metrangolo and Resnati demonstrated the utility of pentafluoroiodobenzene for solid-state structures. Various alkoxystilbazoles have been utilized for nonlinear optics and metallomesogens. Using another finding of Resnati (e.g. N−I complexes form strongly), the group engineered halogen-bonded complexes with iodopentafluorobenzene and 4-alkoxystilbazoles. X-ray crystallography revealed a N−I distance of 2.811(4) Å and the bonding angle to be 168.4°. Similar N−I distances were measured in solid powders. The N−I distance discovered is shorter than the sum of the Van Der Waals radii for nitrogen and iodine (3.53 Å). The single crystal structure of the molecules indicated that no quadrupolar interactions were present. Interestingly, the complexes in Figure 4 were found to be liquid-crystalline.
To test the notion of polarizability involvement in the strength of halogen bonding, bromopentafluorbenzene was used as a Lewis base. Consequently, verification of halogen bond complex formation wasn’t obtained. This finding provides more support for the dependence of halogen bonding on atomic polarizability. Utilizing similar donor-acceptor frameworks, the authors demonstrated that halogen bonding strength in the liquid crystalline state is comparable to the hydrogen-bonded mesogens.
 Applications utilizing properties of conjugated polymers emerged from work done by Heeger, McDiaramid, and Shirakawa
Applications utilizing properties of conjugated polymers emerged from work done by Heeger, McDiaramid, and Shirakawa
with the discovery that polyacetylene
is a conducting, albeit difficult to process material. Since then, work has been done to mimic this conjugated polymer’s backbone (e.g., poly(p-phenylenevinylene)). Conjugated polymers have many practical applications, and are used in devices such as photovoltaic cells, organic light-emitting diode
s, field-effect transistor
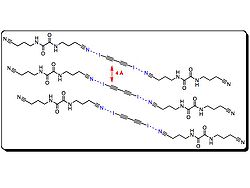
s, and chemical sensors. Goroff et al. prepared ordered poly(diiododiacetylene) (PIDA) via prearrangement of monomer (2) with a halogen bond scaffolding. PIDA is an excellent precursor to other conjugated polymers, as Iodine can be easily transformed. For instance, C−I cleavage is possible electrochemical reduction.
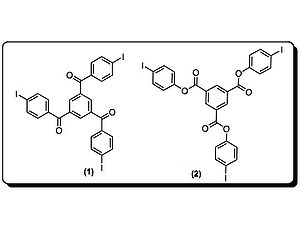
Crystal structures of monomer (2) are disordered materials of varying composition and connectivity. Hosts (3–7) were investigated for their molecular packing, primarily by studying co-crystals of monomer (2) and respective host. Both (3) and (4) pre-organized monomer (2), but steric crowding around the iodines prevented successful topological polymerization of the monomer. Hosts (5–7) utilize hydrogen bonds and halogen bonds to hold monomer (2) at an optimal distance from each other to facilitate polymerization.
In fact, when host 7 was used, polymerization
occurred spontaneously upon isolation of the co-crystals. Crystal structures show the polymer
strands are all parallel to the hydrogen-bonding network, and the host nitriles are each halogen-bonded to iodine atoms. Interestingly, half of the iodine atoms in (1) in the crystal are in close contact to the oxalamide oxygen atoms. Oxygen atoms of host 7 are acting as both hydrogen and halogen bond acceptors.
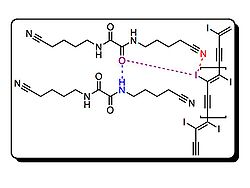
 Porous structures have a variety of uses. Many chemists and material scientists
Porous structures have a variety of uses. Many chemists and material scientists
are working to improve metal-organic frameworks
(MOFs) to store hydrogen to use in cars. These highly organized crystalline inclusion complexes have potential uses in catalysis
and molecular separation devices. Molecular organization is oftentimes controlled via intermolecular forces such as hydrogen bonding. However, utilizing hydrogen bonding often limits the range of pore sizes available due to close packing.
Pigge, et al., utilized halogen bonding interactions between amines, nitrogen heterocycles
, carbonyl
groups, and other organic halides, to construct their porous structures. This is significant because organic crystalline networks mediated by halogen bonds, an interaction significantly weaker than hydrogen bond, are rare.
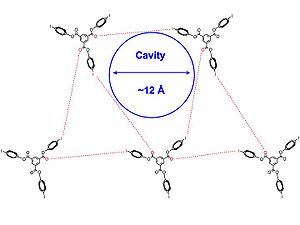
Crystal structures of 1 and 2 [below] were obtained in a variety of solvents, such as dichloromethane
, pyridine
, and benzene
. The authors note that the porous inclusion complexes appear to be mediated in part by unprecedented I-π interactions and by halogen bond between iodine and carbonyl
groups. The crystal structure
[shown below] come together in a triangular array and molecules of 2 are approximately symmetric. Additionally, all of the sets of halogen bonding interactions are not identical, and all of the intermolecular interactions between halogen and halogen bond acceptor slightly exceed the sum of the Van Der Waals radius
, signifying a slightly weaker halogen bond, which leads to more flexibility in the structure. The 2D layers stack parallel to each other to produce channels filled with solvent.
 Solvent interactions are also noted in the formation of the hexagonal structures, especially in pyridine
Solvent interactions are also noted in the formation of the hexagonal structures, especially in pyridine
and chloroform
. Initially, crystals that form these solutions form channeled structures. Over time, new needle-like solvate-free structures form are packed tighter together, and these needles are actually the thermodynamically favored crystal. The authors hope to use this information to better understand the complementary nature of hydrogen bonds and halogen bonds in order to design small molecules predict structures.
(PDB) (July 2004 version), a study by Auffinger and others on single crystals structures with 3 Å resolution or better entered into the PDB revealed that over 100 halogen bonds were found in six halogenated-based nucleic acid structures and sixty-six protein-substrate complexes for halogen-oxygen interactions. Although not as frequent as halogen-oxygen interactions, halogen-nitrogen and halogen-sulfur contacts were identified as well. These scientific findings provide a unique basis for elucidating the role of halogen bonding in biological systems.
On the bio-molecular level, halogen bonding is important for substrate specificity, binding and molecular folding. In the case of protein-ligand interactions, the most common charge-transfer bonds with polarizable halogens involve backbone carbonyls and/or hydroxyl and carboxylate groups of amino acid residues. Typically in DNA
and protein-ligand complexes, the bond distance between Lewis base donor atoms (e.g. O, S, N) and Lewis acid (halogen) is shorter than the sum of their Van der Waals radius. Depending on the structural and chemical environment, halogen bonding interactions can be weak or strong. In the case of some protein-ligand complexes, halogen bonds are energetically and geometrically comparable to that of hydrogen bonding if the donor-acceptor directionality remains consistent. This intermolecular interaction has been shown to be stabilizing and a conformational determinant in protein-ligand and DNA structures.
For molecular recognition and binding, halogen bonding can be significant. An example of this assertion in drug design is the substrate specificity for the binding of IDD 594 to human aldose reductase
. E.I. Howard reported the best resolution for this monomeric enzyme. This biological macromolecule consists of 316 residues, and it reduces aldoses, corticosteroids, and aldehydes. D-sorbitol, a product of the enzymatic conversion of D-glucose, is thought to contribute to the downstream effects of the pathology of diabetes. Hence, inhibiting this enzyme has therapeutic merit.
Aldehyde-based and carboxylate inhibitors are effective but toxic because the functional activity of aldehyde reductase is impaired. Carboxylate and aldehyde
inhibitors were shown to hydrogen bond with Trp 111, Tyr 48, and His 110. The “specificity pocket,” created as a result of inhibitor binding, consists of Leu 300, Ala 299, Phe 122, Thr 113, and Trp 111. For inhibitors to be effective, the key residues of interaction were identified to be Thr 113 and Trp 111. IDD 594 was designed such that the halogen would provide selectivity and be potent. Upon binding, this compound induces a conformational change that causes halogen bonding to occur between the oxygen of the Thr and the bromine of the inhibitor. The bond distance was measured to be 2.973(4) Å. It is this O−Br halogen bond that contributes to the large potency of this inhibitor for human aldose reductase rather than aldehyde reductase.
Halogen
The halogens or halogen elements are a series of nonmetal elements from Group 17 IUPAC Style of the periodic table, comprising fluorine , chlorine , bromine , iodine , and astatine...
atom (Lewis acid) and a Lewis base. Although halogens are involved in other types of bonding (e.g. covalent), halogen bonding specifically refers to when the halogen acts as an electrophilic species.
Bonding
Comparison between hydrogen and halogen bonding:- Hydrogen bonding:
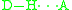
- Halogen bonding:
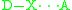
In both cases, D (donor) is the atom, group, or molecule that donates the electron poor species (H or X). H is the hydrogen atom involved in HB, and X is the halogen atom involved in XB. A (acceptor) is the electron rich species.
A parallel relationship can easily be drawn between halogen bonding and hydrogen bonding (HB). In both types of bonding, an electron donor
Electron donor
An electron donor is a chemical entity that donates electrons to another compound. It is a reducing agent that, by virtue of its donating electrons, is itself oxidized in the process....
/electron acceptor
Electron acceptor
An electron acceptor is a chemical entity that accepts electrons transferred to it from another compound. It is an oxidizing agent that, by virtue of its accepting electrons, is itself reduced in the process....
relationship exists. The difference between the two is what species can act as the electron donor/electron acceptor. In hydrogen bonding, a hydrogen atom acts as the electron acceptor and forms a non-covalent interaction by accepting electron density
Electron density
Electron density is the measure of the probability of an electron being present at a specific location.In molecules, regions of electron density are usually found around the atom, and its bonds...
from an electron rich site (electron donor). In halogen bonding, a halogen atom is the electron acceptor. Electron density transfers results in a penetration of the van der Waals
Van der Waals radius
The van der Waals radius, r, of an atom is the radius of an imaginary hard sphere which can be used to model the atom for many purposes. It is named after Johannes Diderik van der Waals, winner of the 1910 Nobel Prize in Physics, as he was the first to recognise that atoms had a finite size and to...
volumes.

Iodine
Iodine is a chemical element with the symbol I and atomic number 53. The name is pronounced , , or . The name is from the , meaning violet or purple, due to the color of elemental iodine vapor....
(I), bromine
Bromine
Bromine ") is a chemical element with the symbol Br, an atomic number of 35, and an atomic mass of 79.904. It is in the halogen element group. The element was isolated independently by two chemists, Carl Jacob Löwig and Antoine Jerome Balard, in 1825–1826...
(Br), chlorine
Chlorine
Chlorine is the chemical element with atomic number 17 and symbol Cl. It is the second lightest halogen, found in the periodic table in group 17. The element forms diatomic molecules under standard conditions, called dichlorine...
(Cl), and sometimes fluorine
Fluorine
Fluorine is the chemical element with atomic number 9, represented by the symbol F. It is the lightest element of the halogen column of the periodic table and has a single stable isotope, fluorine-19. At standard pressure and temperature, fluorine is a pale yellow gas composed of diatomic...
(F). All four halogens are capable of acting as XB donors (as proven through theoretical and experimental data) and follow the general trend: F < Cl < Br < I, with iodine normally forming the strongest interactions.
Dihalogens (I2, Br2, etc.) tend to form strong halogen bonds. The strength and effectiveness of chlorine and fluorine in XB formation depend on the nature of the XB donor. If the halogen is bonded to an electronegative (electron withdrawing) moiety, it is more likely to form stronger halogen bonds.
For example, iodoperfluoroalkanes are well-designed for XB crystal engineering
Crystal engineering
Crystal engineering is the design and synthesis of molecular solid-state structures with desired properties, based on an understanding and exploitation of intermolecular interactions. The two main strategies currently in use for crystal engineering are based on hydrogen bonding and coordination...
. In addition, this is also why F2 can act as a strong XB donor, but fluorocarbon
Fluorocarbon
Fluorocarbons, sometimes referred to as perfluorocarbons or PFCs, are organofluorine compounds that contain only carbon and fluorine bonded together in strong carbon–fluorine bonds. Fluoroalkanes that contain only single bonds are more chemically and thermally stable than alkanes...
s are weak XB donors because the alkyl group connected to the fluorine is not electronegative. In addition, the Lewis base (XB acceptor) tends to be electronegative as well and anions are better XB acceptors than neutral molecules.
Halogen bonds are strong, specific, and directional interactions that give rise to well-defined structures. Halogen bond strengths range from 5–180 kJ/mol. The strength of XB allows it to compete with HB, which are a little bit weaker in strength. Halogen bonds tend to form at 180° angles, which was shown in Odd Hassel
Odd Hassel
Odd Hassel was a Norwegian physical chemist and Nobel Laureate.-Biography:Born in Kristiania, his parents were Ernst Hassel, a gynaecologist, and Mathilde Klaveness. In 1915, he entered the University of Oslo where he studied mathematics, physics and chemistry, and graduated in 1920...
’s studies with bromine and 1,4-dioxane in 1954. Another contributing factor to halogen bond strength comes from the short distance between the halogen (Lewis acid, XB donor) and Lewis base (XB acceptor). The attractive nature of halogen bonds result in the distance between the donor and acceptor to be shorter than the sum of van der Waals radii. The XB interaction becomes stronger as the distance decreases between the halogen and Lewis base.
History
In 1863, Frederick GuthrieFrederick Guthrie
Frederick Guthrie was a British scientific writer and professor. He helped found the Physical Society of London in 1874 and was president of the society from 1884-1886. He believed that science should be based on experimentation rather than discussion...
gave the first report on the ability of halogen atoms to form well-defined adducts with electron donor species. In his experiment, he added I2 to a saturated solution of ammonium nitrate to form NH3I2. When the compound was exposed to air, it spontaneously decomposed into ammonia
Ammonia
Ammonia is a compound of nitrogen and hydrogen with the formula . It is a colourless gas with a characteristic pungent odour. Ammonia contributes significantly to the nutritional needs of terrestrial organisms by serving as a precursor to food and fertilizers. Ammonia, either directly or...
and iodine which allowed Guthrie to conclude that he had formed NH3I2.
In the 1950s, Robert S. Mulliken
Robert S. Mulliken
Robert Sanderson Mulliken was an American physicist and chemist, primarily responsible for the early development of molecular orbital theory, i.e. the elaboration of the molecular orbital method of computing the structure of molecules. Dr. Mulliken received the Nobel Prize for chemistry in 1966...
developed a detailed theory of electron donor-acceptor complexes, classifying them as being outer or inner complexes. Outer complexes were those in which the intermolecular interaction between the electron donor and acceptor were weak and had very little charge transfer. Inner complexes have extensive charge redistribution. Mulliken’s theory has been used to describe the mechanism by which XB formation occurs.

The first X-ray crystallography study from Hassel’s group came in 1954. In the experiment, his group was able to show the structure of bromine 1,4-dioxanate using x-ray diffraction techniques. The experiment revealed that a short intermolecular interaction was present between the oxygen atoms of dioxane and bromine atoms. The O−Br distance in the crystal was measured at 2.71 Å, which indicates a strong interaction between the bromine and oxygen atoms. In addition, the distance is smaller than the sum of the van der Waals radii of oxygen and bromine (3.35 Å). The angle between the O−Br and Br−Br bond is about 180°. This was the first evidence of the typical characteristics found in halogen bond formation and led Hassel to conclude that halogen atoms are directly linked to electron pair donor with a bond direction that coincides with the axes of the orbitals of the lone pairs in the electron pair donor molecule.
In the 1980s continued work was carried out using analytical methods such as infrared spectroscopy
Infrared spectroscopy
Infrared spectroscopy is the spectroscopy that deals with the infrared region of the electromagnetic spectrum, that is light with a longer wavelength and lower frequency than visible light. It covers a range of techniques, mostly based on absorption spectroscopy. As with all spectroscopic...
and Fourier transform spectroscopy
Fourier transform spectroscopy
Fourier transform spectroscopy is a measurement technique whereby spectra are collected based on measurements of the coherence of a radiative source, using time-domain or space-domain measurements of the electromagnetic radiation or other type of radiation....
. These methods allowed the isolation of complexes formed between Lewis bases and halogen molecules for further studies.
Crystal Engineering
Crystal engineeringCrystal engineering
Crystal engineering is the design and synthesis of molecular solid-state structures with desired properties, based on an understanding and exploitation of intermolecular interactions. The two main strategies currently in use for crystal engineering are based on hydrogen bonding and coordination...
is a growing research area that bridges solid-state and supramolecular chemistry. This unique field is interdisciplinary and merges traditional disciplines such as crystallography
Crystallography
Crystallography is the experimental science of the arrangement of atoms in solids. The word "crystallography" derives from the Greek words crystallon = cold drop / frozen drop, with its meaning extending to all solids with some degree of transparency, and grapho = write.Before the development of...
, organic chemistry
Organic chemistry
Organic chemistry is a subdiscipline within chemistry involving the scientific study of the structure, properties, composition, reactions, and preparation of carbon-based compounds, hydrocarbons, and their derivatives...
, and inorganic chemistry
Inorganic chemistry
Inorganic chemistry is the branch of chemistry concerned with the properties and behavior of inorganic compounds. This field covers all chemical compounds except the myriad organic compounds , which are the subjects of organic chemistry...
. In 1971, Schmidt first established the field with a publication on photodimerization in the solid-state. The more recent definition identifies crystal engineering as the utilization of the intermolecular interactions for crystallization and for the development of new substances with different desired physicochemical properties. Before the discovery of halogen bonding, the approach for crystal engineering involved using hydrogen bonding, coordination chemistry and inter-ion interactions for the development of liquid-crystalline and solid-crystalline materials. Furthermore, halogen bonding is employed for the organization of radical cationic salts, fabrication of molecular conductors, and creation of liquid crystal constructs. Since the discovery halogen bonding, new molecular assemblies exist. Due to the unique chemical nature of halogen bonding, this intermolecular interaction serves as an additional tool for the development of crystal engineering.
The first reported use of halogen bonding in liquid crystal formation was by H. Loc Nguyen. In an effort to form liquid crystals, alkoxystilbazoles and pentafluoroiodobenzene were used. Previous studies by Metrangolo and Resnati demonstrated the utility of pentafluoroiodobenzene for solid-state structures. Various alkoxystilbazoles have been utilized for nonlinear optics and metallomesogens. Using another finding of Resnati (e.g. N−I complexes form strongly), the group engineered halogen-bonded complexes with iodopentafluorobenzene and 4-alkoxystilbazoles. X-ray crystallography revealed a N−I distance of 2.811(4) Å and the bonding angle to be 168.4°. Similar N−I distances were measured in solid powders. The N−I distance discovered is shorter than the sum of the Van Der Waals radii for nitrogen and iodine (3.53 Å). The single crystal structure of the molecules indicated that no quadrupolar interactions were present. Interestingly, the complexes in Figure 4 were found to be liquid-crystalline.
To test the notion of polarizability involvement in the strength of halogen bonding, bromopentafluorbenzene was used as a Lewis base. Consequently, verification of halogen bond complex formation wasn’t obtained. This finding provides more support for the dependence of halogen bonding on atomic polarizability. Utilizing similar donor-acceptor frameworks, the authors demonstrated that halogen bonding strength in the liquid crystalline state is comparable to the hydrogen-bonded mesogens.
Preparation of poly(diiododiacetylene)

Hideki Shirakawa
Hideki Shirakawa is a Japanese chemist and winner of the 2000 Nobel Prize in Chemistry for his discovery of conductive polymers together with physics professor Alan J. Heeger and chemistry professor Alan G...
with the discovery that polyacetylene
Polyacetylene
Polyacetylene is an organic polymer with the repeat unit n. The high electrical conductivity discovered for these polymers beginning in the 1960's accelerated interest in the use of organic compounds in microelectronics...
is a conducting, albeit difficult to process material. Since then, work has been done to mimic this conjugated polymer’s backbone (e.g., poly(p-phenylenevinylene)). Conjugated polymers have many practical applications, and are used in devices such as photovoltaic cells, organic light-emitting diode
Organic light-emitting diode
An OLED is a light-emitting diode in which the emissive electroluminescent layer is a film of organic compounds which emit light in response to an electric current. This layer of organic semiconductor material is situated between two electrodes...
s, field-effect transistor
Field-effect transistor
The field-effect transistor is a transistor that relies on an electric field to control the shape and hence the conductivity of a channel of one type of charge carrier in a semiconductor material. FETs are sometimes called unipolar transistors to contrast their single-carrier-type operation with...
s, and chemical sensors. Goroff et al. prepared ordered poly(diiododiacetylene) (PIDA) via prearrangement of monomer (2) with a halogen bond scaffolding. PIDA is an excellent precursor to other conjugated polymers, as Iodine can be easily transformed. For instance, C−I cleavage is possible electrochemical reduction.
Crystal structures of monomer (2) are disordered materials of varying composition and connectivity. Hosts (3–7) were investigated for their molecular packing, primarily by studying co-crystals of monomer (2) and respective host. Both (3) and (4) pre-organized monomer (2), but steric crowding around the iodines prevented successful topological polymerization of the monomer. Hosts (5–7) utilize hydrogen bonds and halogen bonds to hold monomer (2) at an optimal distance from each other to facilitate polymerization.
In fact, when host 7 was used, polymerization
Polymerization
In polymer chemistry, polymerization is a process of reacting monomer molecules together in a chemical reaction to form three-dimensional networks or polymer chains...
occurred spontaneously upon isolation of the co-crystals. Crystal structures show the polymer
Polymer
A polymer is a large molecule composed of repeating structural units. These subunits are typically connected by covalent chemical bonds...
strands are all parallel to the hydrogen-bonding network, and the host nitriles are each halogen-bonded to iodine atoms. Interestingly, half of the iodine atoms in (1) in the crystal are in close contact to the oxalamide oxygen atoms. Oxygen atoms of host 7 are acting as both hydrogen and halogen bond acceptors.


Porous structures

Materials science
Materials science is an interdisciplinary field applying the properties of matter to various areas of science and engineering. This scientific field investigates the relationship between the structure of materials at atomic or molecular scales and their macroscopic properties. It incorporates...
are working to improve metal-organic frameworks
Metal-organic framework
Metal-Organic Frameworks are crystalline compounds consisting of metal ions or clusters coordinated to often rigid organic molecules to form one-, two-, or three-dimensional structures that can be porous. In some cases, the pores are stable to elimination of the guest molecules and can be used for...
(MOFs) to store hydrogen to use in cars. These highly organized crystalline inclusion complexes have potential uses in catalysis
Catalysis
Catalysis is the change in rate of a chemical reaction due to the participation of a substance called a catalyst. Unlike other reagents that participate in the chemical reaction, a catalyst is not consumed by the reaction itself. A catalyst may participate in multiple chemical transformations....
and molecular separation devices. Molecular organization is oftentimes controlled via intermolecular forces such as hydrogen bonding. However, utilizing hydrogen bonding often limits the range of pore sizes available due to close packing.
Pigge, et al., utilized halogen bonding interactions between amines, nitrogen heterocycles
Heterocyclic compound
A heterocyclic compound is a cyclic compound which has atoms of at least two different elements as members of its ring. The counterparts of heterocyclic compounds are homocyclic compounds, the rings of which are made of a single element....
, carbonyl
Carbonyl
In organic chemistry, a carbonyl group is a functional group composed of a carbon atom double-bonded to an oxygen atom: C=O. It is common to several classes of organic compounds, as part of many larger functional groups....
groups, and other organic halides, to construct their porous structures. This is significant because organic crystalline networks mediated by halogen bonds, an interaction significantly weaker than hydrogen bond, are rare.
Crystal structures of 1 and 2 [below] were obtained in a variety of solvents, such as dichloromethane
Dichloromethane
Dichloromethane is an organic compound with the formula CH2Cl2. This colorless, volatile liquid with a moderately sweet aroma is widely used as a solvent. Although it is not miscible with water, it is miscible with many organic solvents...
, pyridine
Pyridine
Pyridine is a basic heterocyclic organic compound with the chemical formula C5H5N. It is structurally related to benzene, with one C-H group replaced by a nitrogen atom...
, and benzene
Benzene
Benzene is an organic chemical compound. It is composed of 6 carbon atoms in a ring, with 1 hydrogen atom attached to each carbon atom, with the molecular formula C6H6....
. The authors note that the porous inclusion complexes appear to be mediated in part by unprecedented I-π interactions and by halogen bond between iodine and carbonyl
Carbonyl
In organic chemistry, a carbonyl group is a functional group composed of a carbon atom double-bonded to an oxygen atom: C=O. It is common to several classes of organic compounds, as part of many larger functional groups....
groups. The crystal structure
Crystal structure
In mineralogy and crystallography, crystal structure is a unique arrangement of atoms or molecules in a crystalline liquid or solid. A crystal structure is composed of a pattern, a set of atoms arranged in a particular way, and a lattice exhibiting long-range order and symmetry...
[shown below] come together in a triangular array and molecules of 2 are approximately symmetric. Additionally, all of the sets of halogen bonding interactions are not identical, and all of the intermolecular interactions between halogen and halogen bond acceptor slightly exceed the sum of the Van Der Waals radius
Van der Waals radius
The van der Waals radius, r, of an atom is the radius of an imaginary hard sphere which can be used to model the atom for many purposes. It is named after Johannes Diderik van der Waals, winner of the 1910 Nobel Prize in Physics, as he was the first to recognise that atoms had a finite size and to...
, signifying a slightly weaker halogen bond, which leads to more flexibility in the structure. The 2D layers stack parallel to each other to produce channels filled with solvent.

Pyridine
Pyridine is a basic heterocyclic organic compound with the chemical formula C5H5N. It is structurally related to benzene, with one C-H group replaced by a nitrogen atom...
and chloroform
Chloroform
Chloroform is an organic compound with formula CHCl3. It is one of the four chloromethanes. The colorless, sweet-smelling, dense liquid is a trihalomethane, and is considered somewhat hazardous...
. Initially, crystals that form these solutions form channeled structures. Over time, new needle-like solvate-free structures form are packed tighter together, and these needles are actually the thermodynamically favored crystal. The authors hope to use this information to better understand the complementary nature of hydrogen bonds and halogen bonds in order to design small molecules predict structures.
Halogen Bonding in Biological Macromolecules
For some time, the significance of halogen bonding to biological macromolecular structure was overlooked. Based on single-crystal structures in the protein data bankProtein Data Bank
The Protein Data Bank is a repository for the 3-D structural data of large biological molecules, such as proteins and nucleic acids....
(PDB) (July 2004 version), a study by Auffinger and others on single crystals structures with 3 Å resolution or better entered into the PDB revealed that over 100 halogen bonds were found in six halogenated-based nucleic acid structures and sixty-six protein-substrate complexes for halogen-oxygen interactions. Although not as frequent as halogen-oxygen interactions, halogen-nitrogen and halogen-sulfur contacts were identified as well. These scientific findings provide a unique basis for elucidating the role of halogen bonding in biological systems.
On the bio-molecular level, halogen bonding is important for substrate specificity, binding and molecular folding. In the case of protein-ligand interactions, the most common charge-transfer bonds with polarizable halogens involve backbone carbonyls and/or hydroxyl and carboxylate groups of amino acid residues. Typically in DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
and protein-ligand complexes, the bond distance between Lewis base donor atoms (e.g. O, S, N) and Lewis acid (halogen) is shorter than the sum of their Van der Waals radius. Depending on the structural and chemical environment, halogen bonding interactions can be weak or strong. In the case of some protein-ligand complexes, halogen bonds are energetically and geometrically comparable to that of hydrogen bonding if the donor-acceptor directionality remains consistent. This intermolecular interaction has been shown to be stabilizing and a conformational determinant in protein-ligand and DNA structures.
For molecular recognition and binding, halogen bonding can be significant. An example of this assertion in drug design is the substrate specificity for the binding of IDD 594 to human aldose reductase
Aldose reductase
Aldose reductase is an NADPH-dependent oxidoreductase that catalyzes the reduction of a variety of aldehydes and carbonyls, including monosaccharides...
. E.I. Howard reported the best resolution for this monomeric enzyme. This biological macromolecule consists of 316 residues, and it reduces aldoses, corticosteroids, and aldehydes. D-sorbitol, a product of the enzymatic conversion of D-glucose, is thought to contribute to the downstream effects of the pathology of diabetes. Hence, inhibiting this enzyme has therapeutic merit.
Aldehyde-based and carboxylate inhibitors are effective but toxic because the functional activity of aldehyde reductase is impaired. Carboxylate and aldehyde
Aldehyde
An aldehyde is an organic compound containing a formyl group. This functional group, with the structure R-CHO, consists of a carbonyl center bonded to hydrogen and an R group....
inhibitors were shown to hydrogen bond with Trp 111, Tyr 48, and His 110. The “specificity pocket,” created as a result of inhibitor binding, consists of Leu 300, Ala 299, Phe 122, Thr 113, and Trp 111. For inhibitors to be effective, the key residues of interaction were identified to be Thr 113 and Trp 111. IDD 594 was designed such that the halogen would provide selectivity and be potent. Upon binding, this compound induces a conformational change that causes halogen bonding to occur between the oxygen of the Thr and the bromine of the inhibitor. The bond distance was measured to be 2.973(4) Å. It is this O−Br halogen bond that contributes to the large potency of this inhibitor for human aldose reductase rather than aldehyde reductase.

