Human evolutionary genetics
Encyclopedia
Human evolutionary genetics studies how one human genome
differs from the other, the evolutionary past that gave rise to it, and its current effects. Differences between genomes have anthropological, medical and forensic implications and applications. Genetic data can provide important insight into human evolution
.
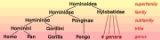
 Biologists classify human
Biologists classify human
s, along with only a few other species
, as great apes (species in the family
Hominidae
). The Hominidae include two distinct species of chimpanzee
(the bonobo
, Pan paniscus, and the common chimpanzee
, Pan troglodytes), two species of gorilla
(the western gorilla
, Gorilla gorilla, and the eastern gorilla
, Gorilla graueri), and two species of orangutan
(the Bornean orangutan
, Pongo pygmaeus, and the Sumatran orangutan
, Pongo abelii).
Apes, in turn, belong to the primate
s order (>400 species). Data from both mitochondrial DNA
(mtDNA) and nuclear DNA
(nDNA) indicates that primates belong to the group of Euarchontoglires
, together with Rodent
ia, Lagomorpha
, Dermoptera, and Scandentia. This is further supported by Alu-like short interspersed nuclear elements (SINEs) which have been found only in members of the Euarchontoglires.
 A phylogenetic tree
A phylogenetic tree
like the one shown above is usually derived from DNA
or protein
sequences
from populations. Often mitochondrial DNA
or Y chromosome
sequences are used to study ancient human demographics. These single-locus
sources of DNA do not recombine
and are almost always inherited from a single parent, with only one known exception in mtDNA (Schwartz and Vissing 2002). Individuals from the various continental groups tend to be more similar to one another than to people from other continents. The tree is rooted in the common ancestor of chimpanzee
s and humans, which is believed to have originated in Africa
. Horizontal distance in the diagram corresponds to two things:
Chimpanzees and humans belong to different genera
, indicated in red. Formation of species
and subspecies
is also indicated, and the formation of races is indicated in the green rectangle to the right (note that only a very rough representation of human phylogeny is given). Note that vertical distances are not meaningful in this representation.
. It has also been shown that the sequence divergence between DNA from humans and chimpanzees varies greatly. For example the sequence divergence varies between 0% to 2.66% between non-coding, non-repetitive genomic regions of humans and chimpanzees. Additionally gene trees, generated by comparative analysis of DNA segments, do not always fit the species tree. Summing up:
from one species (human albumin) induces in the immune system of another species (human, chimpanzee, gorilla and Old World monkey
s). Closely related species should have similar antigens and therefore weaker immunological response to each other's antigens. The immunological response of a species to its own antigens (e.g. human to human) was set to be 1. The ID between humans and gorillas was determined to be 1.09, that between humans and chimpanzees was determined as 1.14. However the distance to six different Old World monkeys was on average 2.46 indicating that the African apes are far closer related to humans than to monkeys. The authors consider the divergence time between Old World monkeys and hominoids to be 30 million years ago (MYA), based on fossil data, and the immunological distance was considered to grow at a constant rate. They concluded that divergence time of humans and the African apes to be roughly ~5 MYA. That was a surprising result. Most scientists at that time thought that humans and great apes diverged much earlier (>15 MYA). The gorilla was, in ID terms, closer to human than to chimpanzees, however the difference was so slight that the trichotomy
could not be resolved with certainty. Later studies based on molecular genetics were able to resolve the trichotomy: chimpanzees are phylogenetically
closer to humans than to gorillas. However, the divergence times estimated later (using much more sophisticated methods in molecular genetics) do not substantially differ from the very first estimate in 1967.
 Current methods to determine divergence times use DNA sequence alignments and molecular clock
Current methods to determine divergence times use DNA sequence alignments and molecular clock
s. Usually the molecular clock is calibrated assuming that the orangutan split from the African apes (including humans) 12-16 MYA. Some studies also include some old world monkeys and set the divergence time of them from hominoids to 25-30 MYA. Both calibration points are based on very little fossil data and have been criticized. If these dates are revised, the divergence times estimated from molecular data will change as well. However, the relative divergence times are unlikely to change. Even if we can't tell absolute divergence times exactly, we can be pretty sure that the divergence time between chimpanzees and humans is about sixfold shorter than between chimpanzees (or humans) and monkeys.
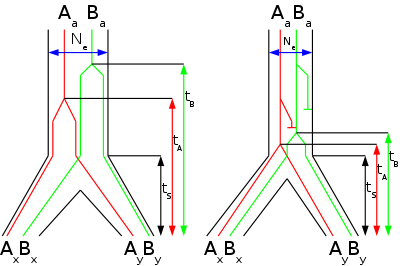
One study (Takahata et al., 1995) used 15 DNA sequence from different regions of the genome from human and chimpanzee and 7 DNA sequences from human, chimpanzee and gorilla. They determined that chimpanzees are more closely related to humans than gorillas. Using various statistical methods, they estimated the divergence time human-chimp to be 4.7 MYA and the divergence time between gorillas and humans (and chimps) to be 7.2 MYA. Additionally they estimated the effective population size
of the common ancestor of humans and chimpanzees to be ~100,000. This was somewhat surprising since the present day effective population size of humans is estimate to be only ~10,000. If true that means that the human lineage would have experienced an immense decrease of its effective population size (and thus genetic diversity) in its evolution. (see Toba catastrophe theory
)
 Another study (Chen & Li, 2001) sequenced 53 non-repetitive, intergenic DNA segments from a human, a chimpanzee, a gorilla, and orangutan. When the DNA sequences were concatenated to a single long sequence, the generated neighbor-joining
Another study (Chen & Li, 2001) sequenced 53 non-repetitive, intergenic DNA segments from a human, a chimpanzee, a gorilla, and orangutan. When the DNA sequences were concatenated to a single long sequence, the generated neighbor-joining
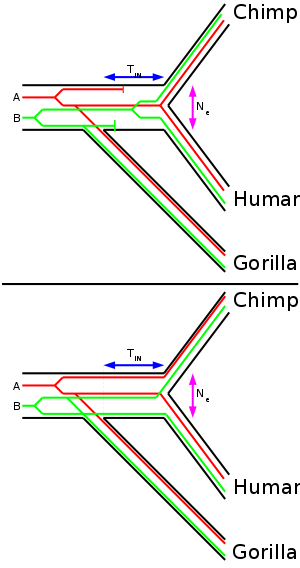
tree supported the Homo-Pan clade with 100% bootstrap (that is that humans and chimpanzees are the closest related species of the four). When three species are fairly closely related to each other (like human, chimpanzee and gorilla), the trees obtained from DNA sequence data may not be congruent with the tree that represents the speciation (species tree). The shorter internodal time span (TIN) the more common are incongruent gene trees. The effective population size (Ne) of the internodal population determines how long genetic lineages are preserved in the population. A higher effective population size causes more incongruent gene trees. Therefore, if the internodal time span is known, the ancestral effective population size of the common ancestor of humans and chimpanzees can be calculated.
When each segment was analyzed individually, 31 supported the Homo-Pan clade, 10 supported the Homo-Gorilla clade, and 12 supported the Pan-Gorilla clade. Using the molecular clock the authors estimated that gorillas split up first 6.2-8.4 MYA and chimpanzees and humans split up 1.6-2.2 million years later (internodal time span) 4.6-6.2 MYA. The internodal time span is useful to estimate the ancestral effective population size of the common ancestor of humans and chimpanzees.
A parsimonious analysis revealed that 24 loci supported the Homo-Pan clade, 7 supported the Homo-Gorilla clade, 2 supported the Pan-Gorilla clade and 20 gave no resolution. Additionally they took 35 protein coding loci from databases. Of these 12 supported the Homo-Pan clade, 3 the Homo-Gorilla clade, 4 the Pan-Gorilla clade and 16 gave no resolution. Therefore only ~70% of the 52 loci that gave a resolution (33 intergenic, 19 protein coding) support the 'correct' species tree. From the fraction of loci which did not support the species tree and the internodal time span they estimated previously, the effective population of the common ancestor of humans and chimpanzees was estimated to be ~52 000 to 96 000. This value is not as high as that from the first study (Takahata), but still much higher than present day effective population size of humans.
A third study (Yang, 2002) used the same dataset that Chen and Li used but estimated the ancestral effective population of 'only' ~12,000 to 21,000, using a different statistical method.
Since mutation rate is relatively constant, roughly one half of these changes occurred in the human lineage. Only a very tiny fraction of those fixed differences gave rise to the different phenotypes of humans and chimpanzees and finding those is a great challenge. The vast majority of the differences are neutral and do not affect the phenotype
.
Molecular evolution may act in different ways, through protein evolution, gene loss, differential gene regulation and RNA evolution. All are thought to have played some part in human evolution.
80 genes were lost in the human lineage after separation from the last common ancestor with the chimpanzee. 36 of those were for olfactory receptors. Genes involved in chemoreception and immune response are overrepresented. Another study estimated that 86 genes had been lost.
was lost in the human lineage. Keratins are a major component of hairs. Humans still have nine functional type I hair keratin genes but the loss of that particular gene may have caused the thinning of human body hair. The gene loss occurred relatively recently in human evolution—less than 240,000 years ago.
gene MYH16 in the human lineage led to smaller masticatory muscles. They estimated that the mutation that led to the inactivation (a two base pair deletion) occurred 2.4 million years ago, predating the appearance of Homo ergaster
/erectus in Africa. The period that followed was marked by a strong increase in cranial capacity
, promoting speculation that the loss of the gene may have removed an evolutionary constraint on brain size in the genus Homo.
Another estimate for the loss of the MYH16 gene is 5.3 million years ago, long before Homo appeared.
s (SDs or LCRs) have had roles in creating new primate genes and shaping human genetic variation.
are areas of the genome that differ between humans and chimpanzees to a greater extent than can be explained by genetic drift over the time since the two species shared a common ancestor. These regions show signs of being subject to natural selection, leading to the evolution of distinctly human traits. Two examples are HAR1F
, which is believed to be related to brain development and HAR2 (a.k.a HACNS1) that may have played a role in the development of the opposable thumb
.
It has also been hypothesized that much of the difference between humans and chimpanzees is attributable to the regulation of gene expression
rather than differences in the genes themselves. Analyses of conserved non-coding sequence
s, which often contain functional and thus positively selected regulatory regions, address this possibility.
genome in May 2010. The results indicate some breeding between humans and Neanderthals as the genomes of non-African humans have 1-4% more in common with Neanderthals than do the genomes of subsaharan Africans. Neanderthals and most humans share a lactose-intolerant variant of the lactase
gene that encodes an enzyme that is unable to break down lactose in milk after weaning. Humans and Neanderthals also share the FOXP2
gene variant associated with brain development and with speech in humans, indicating that Neanderthals may have been able to speak. Chimps have two amino acid differences in FOXP2 compared with human and Neanderthal FOXP2.
s (insertions/deletions) accounted for many fewer differences (15% as many), but contributed ~1.5% of unique sequence to each genome, since each insertion or deletion can involve anywhere from one base to millions of bases . A companion paper examined segmental duplication
s in the two genomes , whose insertion and deletion into the genome account for much of the indel sequence. They found that a total of 2.7% of euchromatic sequence had been differentially duplicated in one or the other lineage.
The sequence divergence has generally the following pattern: Human-Chimp < Human-Gorilla << Human-Orangutan, highlighting the close kinship between humans and the African apes. Alu elements
diverge quickly due to their high frequency of CpG
dinucleotides which mutate roughly 10 times more often than the average nucleotide in the genome. The mutation rate is higher in the male germ line, therefore the divergence in the Y chromosome
—which is inherited solely from the father—is higher than in autosomes. The X chromosome
is inherited twice as often through the female germ line as through the male germ line and therefore shows slightly lower sequence divergence. The sequence divergence of the Xq13.3 region is surprisingly low between humans and chimpanzees.
Mutations altering the amino acid sequence of proteins (Ka) are the least common. In fact ~29% of all orthologous proteins are identical between human and chimpanzee. The typical protein differs by only two amino acids.
The measures of sequence divergence shown in the table only take the substitutional differences, for example from an A (adenine
) to a G (guanine
), into account. DNA sequences may however also differ by insertions and deletions (indel
s) of bases. These are usually stripped from the alignments before the calculation of sequence divergence is performed.
 Molecular biologists starting with Wesley Brown
Molecular biologists starting with Wesley Brown
on mtDNA and Allan Wilson
on mtDNA have produced observations relevant to human evolution.
(mtMRCA)) of all extant humans has become known as Mitochondrial Eve
. The observation that the mtMRCA is the direct matrilineal ancestor of all living humans does not mean either that she was the first anatomically modern human, nor that no other female humans lived concurrently with her. Other women would have lived at the same time and passed nuclear genes down to living humans, but their mitochondrial lineages were lost over time. This could be due to random events such as producing only male children.
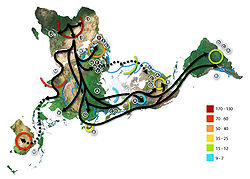
The broad study of African genetic diversity headed by Sarah Tishkoff found the San people
to express the greatest genetic diversity among the 113 distinct populations sampled, making them one of 14 "ancestral population clusters". The research also located the origin of modern human migration in south-western Africa, near the coastal border of Namibia
and Angola
.
is much larger than mtDNA, and is relatively homogeneous; therefore it has taken much longer to find distinct lineages and to analyse them. Conversely, because the Y chromosome is so large by comparison, it holds more genetic information. Y chromosome studies show similar findings to those made with mtDNA. The estimate for the age of the ancestral Y chromosome for all extant Y chromosomes is given at about 70,000 years ago and is also placed in Africa; the individual who contributed this Y chromosomal heritage is sometimes referred to as Y chromosome Adam. The difference in dates between Y chromosome Adam and mitochondrial Eve is usually attributed to a higher extinction rate for Y chromosomes due to greater differential reproductive success between individual men, which means that a small number of very successful men may produce many children, while a larger number of less successful men will produce far fewer children.
Human genome
The human genome is the genome of Homo sapiens, which is stored on 23 chromosome pairs plus the small mitochondrial DNA. 22 of the 23 chromosomes are autosomal chromosome pairs, while the remaining pair is sex-determining...
differs from the other, the evolutionary past that gave rise to it, and its current effects. Differences between genomes have anthropological, medical and forensic implications and applications. Genetic data can provide important insight into human evolution
Human evolution
Human evolution refers to the evolutionary history of the genus Homo, including the emergence of Homo sapiens as a distinct species and as a unique category of hominids and mammals...
.
Origin of apes

Human
Humans are the only living species in the Homo genus...
s, along with only a few other species
Species
In biology, a species is one of the basic units of biological classification and a taxonomic rank. A species is often defined as a group of organisms capable of interbreeding and producing fertile offspring. While in many cases this definition is adequate, more precise or differing measures are...
, as great apes (species in the family
Family (biology)
In biological classification, family is* a taxonomic rank. Other well-known ranks are life, domain, kingdom, phylum, class, order, genus, and species, with family fitting between order and genus. As for the other well-known ranks, there is the option of an immediately lower rank, indicated by the...
Hominidae
Hominidae
The Hominidae or include them .), as the term is used here, form a taxonomic family, including four extant genera: chimpanzees , gorillas , humans , and orangutans ....
). The Hominidae include two distinct species of chimpanzee
Chimpanzee
Chimpanzee, sometimes colloquially chimp, is the common name for the two extant species of ape in the genus Pan. The Congo River forms the boundary between the native habitat of the two species:...
(the bonobo
Bonobo
The bonobo , Pan paniscus, previously called the pygmy chimpanzee and less often, the dwarf or gracile chimpanzee, is a great ape and one of the two species making up the genus Pan. The other species in genus Pan is Pan troglodytes, or the common chimpanzee...
, Pan paniscus, and the common chimpanzee
Common Chimpanzee
The common chimpanzee , also known as the robust chimpanzee, is a great ape. Colloquially, the common chimpanzee is often called the chimpanzee , though technically this term refers to both species in the genus Pan: the common chimpanzee and the closely related bonobo, formerly called the pygmy...
, Pan troglodytes), two species of gorilla
Gorilla
Gorillas are the largest extant species of primates. They are ground-dwelling, predominantly herbivorous apes that inhabit the forests of central Africa. Gorillas are divided into two species and either four or five subspecies...
(the western gorilla
Western Gorilla
The western gorilla is a great ape and the most populous species of the genus Gorilla.-Taxonomy:Nearly all of the individuals of this taxon belong to the western lowland gorilla subspecies whose population is approximately 95,000 individuals...
, Gorilla gorilla, and the eastern gorilla
Eastern Gorilla
The eastern gorilla is a species of the genus Gorilla and the largest living primate. At present, the species is subdivided into two subspecies. The eastern lowland gorilla is the most populous, at about 5,000 individuals. The mountain gorilla has only about 700 individuals...
, Gorilla graueri), and two species of orangutan
Orangutan
Orangutans are the only exclusively Asian genus of extant great ape. The largest living arboreal animals, they have proportionally longer arms than the other, more terrestrial, great apes. They are among the most intelligent primates and use a variety of sophisticated tools, also making sleeping...
(the Bornean orangutan
Bornean Orangutan
The Bornean orangutan, Pongo pygmaeus, is a species of orangutan native to the island of Borneo. Together with the slightly smaller Sumatran orangutan, it belongs to the only genus of great apes native to Asia....
, Pongo pygmaeus, and the Sumatran orangutan
Sumatran Orangutan
The Sumatran orangutan is one of the two species of orangutans. Found only on the island of Sumatra, in Indonesia, it is rarer and smaller than the Bornean orangutan. The Sumatran orangutan grows to about tall and in males...
, Pongo abelii).
Apes, in turn, belong to the primate
Primate
A primate is a mammal of the order Primates , which contains prosimians and simians. Primates arose from ancestors that lived in the trees of tropical forests; many primate characteristics represent adaptations to life in this challenging three-dimensional environment...
s order (>400 species). Data from both mitochondrial DNA
Mitochondrial DNA
Mitochondrial DNA is the DNA located in organelles called mitochondria, structures within eukaryotic cells that convert the chemical energy from food into a form that cells can use, adenosine triphosphate...
(mtDNA) and nuclear DNA
Nuclear DNA
Nuclear DNA, nuclear deoxyribonucleic acid , is DNA contained within a nucleus of eukaryotic organisms. In mammals and vertebrates, nuclear DNA encodes more of the genome than the mitochondrial DNA and is composed of information inherited from two parents, one male, and one female, rather than...
(nDNA) indicates that primates belong to the group of Euarchontoglires
Euarchontoglires
Euarchontoglires is a clade of mammals, the living members of which are rodents, lagomorphs, treeshrews, colugos and primates .-Evolutionary relationships:...
, together with Rodent
Rodent
Rodentia is an order of mammals also known as rodents, characterised by two continuously growing incisors in the upper and lower jaws which must be kept short by gnawing....
ia, Lagomorpha
Lagomorpha
The lagomorphs are the members of the taxonomic order Lagomorpha, of which there are two living families, the Leporidae , and the Ochotonidae...
, Dermoptera, and Scandentia. This is further supported by Alu-like short interspersed nuclear elements (SINEs) which have been found only in members of the Euarchontoglires.
Cladistics

Phylogenetic tree
A phylogenetic tree or evolutionary tree is a branching diagram or "tree" showing the inferred evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical and/or genetic characteristics...
like the one shown above is usually derived from DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
or protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
sequences
DNA sequence
The sequence or primary structure of a nucleic acid is the composition of atoms that make up the nucleic acid and the chemical bonds that bond those atoms. Because nucleic acids, such as DNA and RNA, are unbranched polymers, this specification is equivalent to specifying the sequence of...
from populations. Often mitochondrial DNA
Mitochondrial DNA
Mitochondrial DNA is the DNA located in organelles called mitochondria, structures within eukaryotic cells that convert the chemical energy from food into a form that cells can use, adenosine triphosphate...
or Y chromosome
Y-chromosomal Adam
In human genetics, Y-chromosomal Adam is the theoretical most recent common ancestor from whom all living people are descended patrilineally . Many studies report that Y-chromosomal Adam lived as early as around 142,000 years ago and possibly as recently as 60,000 years ago...
sequences are used to study ancient human demographics. These single-locus
Locus (genetics)
In the fields of genetics and genetic computation, a locus is the specific location of a gene or DNA sequence on a chromosome. A variant of the DNA sequence at a given locus is called an allele. The ordered list of loci known for a particular genome is called a genetic map...
sources of DNA do not recombine
Genetic recombination
Genetic recombination is a process by which a molecule of nucleic acid is broken and then joined to a different one. Recombination can occur between similar molecules of DNA, as in homologous recombination, or dissimilar molecules, as in non-homologous end joining. Recombination is a common method...
and are almost always inherited from a single parent, with only one known exception in mtDNA (Schwartz and Vissing 2002). Individuals from the various continental groups tend to be more similar to one another than to people from other continents. The tree is rooted in the common ancestor of chimpanzee
Chimpanzee
Chimpanzee, sometimes colloquially chimp, is the common name for the two extant species of ape in the genus Pan. The Congo River forms the boundary between the native habitat of the two species:...
s and humans, which is believed to have originated in Africa
Africa
Africa is the world's second largest and second most populous continent, after Asia. At about 30.2 million km² including adjacent islands, it covers 6% of the Earth's total surface area and 20.4% of the total land area...
. Horizontal distance in the diagram corresponds to two things:
- Genetic distance. Given below the diagram, the genetic difference between humans and chimps is less than 2%, or 20 times larger than the variation among modern humans.
- Temporal remoteness of the most recent common ancestor. Rough estimates are given above the diagram, in millions of years. The mitochondrial most recent common ancestor of modern humans lived roughly 200,000 years ago, latest common ancestors of humans and chimps between four and seven million years ago.
Chimpanzees and humans belong to different genera
Genus
In biology, a genus is a low-level taxonomic rank used in the biological classification of living and fossil organisms, which is an example of definition by genus and differentia...
, indicated in red. Formation of species
Species
In biology, a species is one of the basic units of biological classification and a taxonomic rank. A species is often defined as a group of organisms capable of interbreeding and producing fertile offspring. While in many cases this definition is adequate, more precise or differing measures are...
and subspecies
Subspecies
Subspecies in biological classification, is either a taxonomic rank subordinate to species, ora taxonomic unit in that rank . A subspecies cannot be recognized in isolation: a species will either be recognized as having no subspecies at all or two or more, never just one...
is also indicated, and the formation of races is indicated in the green rectangle to the right (note that only a very rough representation of human phylogeny is given). Note that vertical distances are not meaningful in this representation.
Speciation of humans and the African apes
The separation of humans from their closest relatives, the African apes (chimpanzees and gorillas), has been studied extensively for more than a century. Five major questions have been addressed:- Which apes are our closest ancestors?
- When did the separations occur?
- What was the effective population sizeEffective population sizeIn population genetics, the concept of effective population size Ne was introduced by the American geneticist Sewall Wright, who wrote two landmark papers on it...
of the common ancestor before the split? - Are there traces of population structure (subpopulations) preceding the speciation or partial admixture succeeding it?
- What were the specific events (including fusion of chromosomes 2a and 2b) prior to and subsequent to the separation?
General observations
As discussed before, different parts of the genome show different sequence divergence between different hominoidsApe
Apes are Old World anthropoid mammals, more specifically a clade of tailless catarrhine primates, belonging to the biological superfamily Hominoidea. The apes are native to Africa and South-east Asia, although in relatively recent times humans have spread all over the world...
. It has also been shown that the sequence divergence between DNA from humans and chimpanzees varies greatly. For example the sequence divergence varies between 0% to 2.66% between non-coding, non-repetitive genomic regions of humans and chimpanzees. Additionally gene trees, generated by comparative analysis of DNA segments, do not always fit the species tree. Summing up:
- The sequence divergence varies significantly between humans, chimpanzees and gorillas.
- For most DNA sequences, humans and chimpanzees appear to be most closely related, but some point to a human-gorilla or chimpanzee-gorilla cladeCladeA clade is a group consisting of a species and all its descendants. In the terms of biological systematics, a clade is a single "branch" on the "tree of life". The idea that such a "natural group" of organisms should be grouped together and given a taxonomic name is central to biological...
. - The human genome has been sequenced, as well as the chimpanzee genome. Humans have 23 pairs of chromosomes, while chimpanzees, gorillas, and orangutans have 24. Human chromosome 2Chromosome 2 (human)Chromosome 2 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 2 is the second largest human chromosome, spanning more than 237 million base pairs and representing almost 8% of the total DNA in cells.Identifying genes on each...
is a fusion between two chromosomes that remained separate in the other primates.
Divergence times
The divergence time of humans from other apes is of great interest. One of the first molecular studies, published in 1967 measured immunological distances (IDs) between different primates. Basically the study measured the strength of immunological response that an antigenAntigen
An antigen is a foreign molecule that, when introduced into the body, triggers the production of an antibody by the immune system. The immune system will then kill or neutralize the antigen that is recognized as a foreign and potentially harmful invader. These invaders can be molecules such as...
from one species (human albumin) induces in the immune system of another species (human, chimpanzee, gorilla and Old World monkey
Old World monkey
The Old World monkeys or Cercopithecidae are a group of primates, falling in the superfamily Cercopithecoidea in the clade Catarrhini. The Old World monkeys are native to Africa and Asia today, inhabiting a range of environments from tropical rain forest to savanna, shrubland and mountainous...
s). Closely related species should have similar antigens and therefore weaker immunological response to each other's antigens. The immunological response of a species to its own antigens (e.g. human to human) was set to be 1. The ID between humans and gorillas was determined to be 1.09, that between humans and chimpanzees was determined as 1.14. However the distance to six different Old World monkeys was on average 2.46 indicating that the African apes are far closer related to humans than to monkeys. The authors consider the divergence time between Old World monkeys and hominoids to be 30 million years ago (MYA), based on fossil data, and the immunological distance was considered to grow at a constant rate. They concluded that divergence time of humans and the African apes to be roughly ~5 MYA. That was a surprising result. Most scientists at that time thought that humans and great apes diverged much earlier (>15 MYA). The gorilla was, in ID terms, closer to human than to chimpanzees, however the difference was so slight that the trichotomy
Trichotomy
In mathematics, the Law of Trichotomy states that every real number is either positive, negative, or zero. More generally, trichotomy is the property of an order relation...
could not be resolved with certainty. Later studies based on molecular genetics were able to resolve the trichotomy: chimpanzees are phylogenetically
Phylogenetics
In biology, phylogenetics is the study of evolutionary relatedness among groups of organisms , which is discovered through molecular sequencing data and morphological data matrices...
closer to humans than to gorillas. However, the divergence times estimated later (using much more sophisticated methods in molecular genetics) do not substantially differ from the very first estimate in 1967.
Divergence times and ancestral effective population size

Molecular clock
The molecular clock is a technique in molecular evolution that uses fossil constraints and rates of molecular change to deduce the time in geologic history when two species or other taxa diverged. It is used to estimate the time of occurrence of events called speciation or radiation...
s. Usually the molecular clock is calibrated assuming that the orangutan split from the African apes (including humans) 12-16 MYA. Some studies also include some old world monkeys and set the divergence time of them from hominoids to 25-30 MYA. Both calibration points are based on very little fossil data and have been criticized. If these dates are revised, the divergence times estimated from molecular data will change as well. However, the relative divergence times are unlikely to change. Even if we can't tell absolute divergence times exactly, we can be pretty sure that the divergence time between chimpanzees and humans is about sixfold shorter than between chimpanzees (or humans) and monkeys.
One study (Takahata et al., 1995) used 15 DNA sequence from different regions of the genome from human and chimpanzee and 7 DNA sequences from human, chimpanzee and gorilla. They determined that chimpanzees are more closely related to humans than gorillas. Using various statistical methods, they estimated the divergence time human-chimp to be 4.7 MYA and the divergence time between gorillas and humans (and chimps) to be 7.2 MYA. Additionally they estimated the effective population size
Effective population size
In population genetics, the concept of effective population size Ne was introduced by the American geneticist Sewall Wright, who wrote two landmark papers on it...
of the common ancestor of humans and chimpanzees to be ~100,000. This was somewhat surprising since the present day effective population size of humans is estimate to be only ~10,000. If true that means that the human lineage would have experienced an immense decrease of its effective population size (and thus genetic diversity) in its evolution. (see Toba catastrophe theory
Toba catastrophe theory
The Toba supereruption was a supervolcanic eruption that occurred some time between 69,000 and 77,000 years ago at Lake Toba . It is recognized as one of the Earth's largest known eruptions...
)

Neighbor-joining
In bioinformatics, neighbor joining is a bottom-up clustering method for the creation of phenetic trees , created by Naruya Saitou and Masatoshi Nei...
tree supported the Homo-Pan clade with 100% bootstrap (that is that humans and chimpanzees are the closest related species of the four). When three species are fairly closely related to each other (like human, chimpanzee and gorilla), the trees obtained from DNA sequence data may not be congruent with the tree that represents the speciation (species tree). The shorter internodal time span (TIN) the more common are incongruent gene trees. The effective population size (Ne) of the internodal population determines how long genetic lineages are preserved in the population. A higher effective population size causes more incongruent gene trees. Therefore, if the internodal time span is known, the ancestral effective population size of the common ancestor of humans and chimpanzees can be calculated.
When each segment was analyzed individually, 31 supported the Homo-Pan clade, 10 supported the Homo-Gorilla clade, and 12 supported the Pan-Gorilla clade. Using the molecular clock the authors estimated that gorillas split up first 6.2-8.4 MYA and chimpanzees and humans split up 1.6-2.2 million years later (internodal time span) 4.6-6.2 MYA. The internodal time span is useful to estimate the ancestral effective population size of the common ancestor of humans and chimpanzees.
A parsimonious analysis revealed that 24 loci supported the Homo-Pan clade, 7 supported the Homo-Gorilla clade, 2 supported the Pan-Gorilla clade and 20 gave no resolution. Additionally they took 35 protein coding loci from databases. Of these 12 supported the Homo-Pan clade, 3 the Homo-Gorilla clade, 4 the Pan-Gorilla clade and 16 gave no resolution. Therefore only ~70% of the 52 loci that gave a resolution (33 intergenic, 19 protein coding) support the 'correct' species tree. From the fraction of loci which did not support the species tree and the internodal time span they estimated previously, the effective population of the common ancestor of humans and chimpanzees was estimated to be ~52 000 to 96 000. This value is not as high as that from the first study (Takahata), but still much higher than present day effective population size of humans.
A third study (Yang, 2002) used the same dataset that Chen and Li used but estimated the ancestral effective population of 'only' ~12,000 to 21,000, using a different statistical method.
Genetic differences between humans and other great apes
The alignable sequences within genomes of humans and chimpanzees differ by about 35 million single-nucleotide substitutions. Additionally about 3% of the complete genomes differ by deletions, insertions and duplications.Since mutation rate is relatively constant, roughly one half of these changes occurred in the human lineage. Only a very tiny fraction of those fixed differences gave rise to the different phenotypes of humans and chimpanzees and finding those is a great challenge. The vast majority of the differences are neutral and do not affect the phenotype
Phenotype
A phenotype is an organism's observable characteristics or traits: such as its morphology, development, biochemical or physiological properties, behavior, and products of behavior...
.
Molecular evolution may act in different ways, through protein evolution, gene loss, differential gene regulation and RNA evolution. All are thought to have played some part in human evolution.
Gene loss
Many different mutations can inactivate a gene, but few will change its function in a specific way. Inactivation mutations will therefore be readily available for selection to act on. Gene loss could thus be a common mechanism of evolutionary adaptation (the "less-is-more" hypothesis).80 genes were lost in the human lineage after separation from the last common ancestor with the chimpanzee. 36 of those were for olfactory receptors. Genes involved in chemoreception and immune response are overrepresented. Another study estimated that 86 genes had been lost.
Hair keratin gene KRTHAP1
A gene for type I hair keratinKeratin
Keratin refers to a family of fibrous structural proteins. Keratin is the key of structural material making up the outer layer of human skin. It is also the key structural component of hair and nails...
was lost in the human lineage. Keratins are a major component of hairs. Humans still have nine functional type I hair keratin genes but the loss of that particular gene may have caused the thinning of human body hair. The gene loss occurred relatively recently in human evolution—less than 240,000 years ago.
Myosin gene MYH16
Stedman et al. (2004) stated that the loss of the sarcomeric myosinMyosin
Myosins comprise a family of ATP-dependent motor proteins and are best known for their role in muscle contraction and their involvement in a wide range of other eukaryotic motility processes. They are responsible for actin-based motility. The term was originally used to describe a group of similar...
gene MYH16 in the human lineage led to smaller masticatory muscles. They estimated that the mutation that led to the inactivation (a two base pair deletion) occurred 2.4 million years ago, predating the appearance of Homo ergaster
Homo ergaster
Homo ergaster is an extinct chronospecies of Homo that lived in eastern and southern Africa during the early Pleistocene, about 2.5–1.7 million years ago.There is still disagreement on the subject of the classification, ancestry, and progeny of H...
/erectus in Africa. The period that followed was marked by a strong increase in cranial capacity
Cranial capacity
Cranial capacity is a measure of the volume of the interior of the cranium of those vertebrates who have both a cranium and a brain. The most commonly used unit of measure is the cubic centimetre or cc...
, promoting speculation that the loss of the gene may have removed an evolutionary constraint on brain size in the genus Homo.
Another estimate for the loss of the MYH16 gene is 5.3 million years ago, long before Homo appeared.
Other
- CASPASE12Caspase 12Caspase 12 is an enzyme known as a cysteine protease. It belongs to a family of enzymes called caspases that cleave their substrates at C-terminal aspartic acid residues...
, a cysteinyl aspartate proteinase. The loss of this gene is speculated to have reduced the lethality of bacterial infection in humans.
Gene addition
Segmental duplicationSegmental duplication
Segmental duplications are segments of DNA with near-identical sequence.Segmental duplications give rise to low copy repeats and are believed to have played a role in creating new primate genes as reflected in human genetic variation...
s (SDs or LCRs) have had roles in creating new primate genes and shaping human genetic variation.
Selection pressures
Human accelerated regionsHuman accelerated regions
Human accelerated regions , first described in August 2006, are a set of 49 segments of the human genome which are conserved throughout vertebrate evolution but are strikingly different in humans. They are named HAR1 through HAR49 according to their degree of difference between humans and chimpanzees...
are areas of the genome that differ between humans and chimpanzees to a greater extent than can be explained by genetic drift over the time since the two species shared a common ancestor. These regions show signs of being subject to natural selection, leading to the evolution of distinctly human traits. Two examples are HAR1F
HAR1F
In molecular biology, Human accelerated region 1 is a segment of the human genome found on the long arm of chromosome 20. It is a Human accelerated region...
, which is believed to be related to brain development and HAR2 (a.k.a HACNS1) that may have played a role in the development of the opposable thumb
Thumb
The thumb is the first digit of the hand. When a person is standing in the medical anatomical position , the thumb is the lateral-most digit...
.
It has also been hypothesized that much of the difference between humans and chimpanzees is attributable to the regulation of gene expression
Regulation of gene expression
Gene modulation redirects here. For information on therapeutic regulation of gene expression, see therapeutic gene modulation.Regulation of gene expression includes the processes that cells and viruses use to regulate the way that the information in genes is turned into gene products...
rather than differences in the genes themselves. Analyses of conserved non-coding sequence
Conserved non-coding sequence
A conserved non-coding sequence is a DNA sequence of noncoding DNA that is evolutionarily conserved. These sequences are of interest for their potential to regulate gene production....
s, which often contain functional and thus positively selected regulatory regions, address this possibility.
Genetic differences between humans and Neanderthals
An international group of scientists completed a draft sequence of the NeanderthalNeanderthal
The Neanderthal is an extinct member of the Homo genus known from Pleistocene specimens found in Europe and parts of western and central Asia...
genome in May 2010. The results indicate some breeding between humans and Neanderthals as the genomes of non-African humans have 1-4% more in common with Neanderthals than do the genomes of subsaharan Africans. Neanderthals and most humans share a lactose-intolerant variant of the lactase
Lactase
Lactase , a part of the β-galactosidase family of enzymes, is a glycoside hydrolase involved in the hydrolysis of the disaccharide lactose into constituent galactose and glucose monomers...
gene that encodes an enzyme that is unable to break down lactose in milk after weaning. Humans and Neanderthals also share the FOXP2
FOXP2
Forkhead box protein P2 also known as FOXP2 is a protein that in humans is encoded by the FOXP2 gene, located on human chromosome 7 . FOXP2 orthologs have also been identified in all mammals for which complete genome data are available...
gene variant associated with brain development and with speech in humans, indicating that Neanderthals may have been able to speak. Chimps have two amino acid differences in FOXP2 compared with human and Neanderthal FOXP2.
Sequence divergence between humans and apes
When the draft sequence of the common chimpanzee (Pan troglodytes) genome was published in the summer 2005, 2400 million bases (of ~3160 million bases) were sequenced and assembled well enough to be compared to the human genome. 1.23% of this sequenced differed by single-base substitutions. Of this, 1.06% or less was thought to represent fixed differences between the species, with the rest being variant sites in humans or chimpanzees. Another type of difference, called indelIndel
Indel is a molecular biology term that has different definitions in different fields:*In evolutionary studies, indel is used to mean an insertion or a deletion and indels simply refers to the mutation class that includes both insertions, deletions, and the combination thereof, including insertion...
s (insertions/deletions) accounted for many fewer differences (15% as many), but contributed ~1.5% of unique sequence to each genome, since each insertion or deletion can involve anywhere from one base to millions of bases . A companion paper examined segmental duplication
Segmental duplication
Segmental duplications are segments of DNA with near-identical sequence.Segmental duplications give rise to low copy repeats and are believed to have played a role in creating new primate genes as reflected in human genetic variation...
s in the two genomes , whose insertion and deletion into the genome account for much of the indel sequence. They found that a total of 2.7% of euchromatic sequence had been differentially duplicated in one or the other lineage.
| Locus | Human-Chimp | Human-Gorilla | Human-Orangutan |
|---|---|---|---|
| Alu elements | 2 | - | - |
| Non-coding (Chr. Y) | 1.68 ± 0.19 | 2.33 ± 0.2 | 5.63 ± 0.35 |
| Pseudogenes (autosomal) | 1.64 ± 0.10 | 1.87 ± 0.11 | - |
| Pseudogenes (Chr. X) | 1.47 ± 0.17 | - | - |
| Noncoding (autosomal) | 1.24 ± 0.07 | 1.62 ± 0.08 | 3.08 ± 0.11 |
| Genes (Ks) | 1.11 | 1.48 | 2.98 |
| Introns | 0.93 ± 0.08 | 1.23 ± 0.09 | - |
| Xq13.3 | 0.92 ± 0.10 | 1.42 ± 0.12 | 3.00 ± 0.18 |
| Subtotal for X chromosome | 1.16 ± 0.07 | 1.47 ± 0.08 | - |
| Genes (Ka) | 0.8 | 0.93 | 1.96 |
The sequence divergence has generally the following pattern: Human-Chimp < Human-Gorilla << Human-Orangutan, highlighting the close kinship between humans and the African apes. Alu elements
Alu sequence
An Alu element is a short stretch of DNA originally characterized by the action of the Alu restriction endonuclease. Alu elements of different kinds occur in large numbers in primate genomes. In fact, Alu elements are the most abundant mobile elements in the human genome. They are derived from the...
diverge quickly due to their high frequency of CpG
CpG site
CpG sites or CG sites are regions of DNA where a cytosine nucleotide occurs next to a guanine nucleotide in the linear sequence of bases along its length. "CpG" is shorthand for "—C—phosphate—G—", that is, cytosine and guanine separated by only one phosphate; phosphate links any two nucleosides...
dinucleotides which mutate roughly 10 times more often than the average nucleotide in the genome. The mutation rate is higher in the male germ line, therefore the divergence in the Y chromosome
Y chromosome
The Y chromosome is one of the two sex-determining chromosomes in most mammals, including humans. In mammals, it contains the gene SRY, which triggers testis development if present. The human Y chromosome is composed of about 60 million base pairs...
—which is inherited solely from the father—is higher than in autosomes. The X chromosome
X chromosome
The X chromosome is one of the two sex-determining chromosomes in many animal species, including mammals and is common in both males and females. It is a part of the XY sex-determination system and X0 sex-determination system...
is inherited twice as often through the female germ line as through the male germ line and therefore shows slightly lower sequence divergence. The sequence divergence of the Xq13.3 region is surprisingly low between humans and chimpanzees.
Mutations altering the amino acid sequence of proteins (Ka) are the least common. In fact ~29% of all orthologous proteins are identical between human and chimpanzee. The typical protein differs by only two amino acids.
The measures of sequence divergence shown in the table only take the substitutional differences, for example from an A (adenine
Adenine
Adenine is a nucleobase with a variety of roles in biochemistry including cellular respiration, in the form of both the energy-rich adenosine triphosphate and the cofactors nicotinamide adenine dinucleotide and flavin adenine dinucleotide , and protein synthesis, as a chemical component of DNA...
) to a G (guanine
Guanine
Guanine is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine . In DNA, guanine is paired with cytosine. With the formula C5H5N5O, guanine is a derivative of purine, consisting of a fused pyrimidine-imidazole ring system with...
), into account. DNA sequences may however also differ by insertions and deletions (indel
Indel
Indel is a molecular biology term that has different definitions in different fields:*In evolutionary studies, indel is used to mean an insertion or a deletion and indels simply refers to the mutation class that includes both insertions, deletions, and the combination thereof, including insertion...
s) of bases. These are usually stripped from the alignments before the calculation of sequence divergence is performed.
Modern humans

Wesley Brown
Wesley Brown may refer to:*Wes Brown, English football player*Wesley A. Brown, first African-American to graduate from the U.S. Naval Academy*Wesley E. Brown, U.S. District Court judge...
on mtDNA and Allan Wilson
Allan Wilson
Allan Charles Wilson was a pioneer in the use of molecular approaches to understand evolutionary change and reconstruct phylogenies, and a contributor to the study of human evolution. He was one of the most controversial figures in post-war biology; his work attracted a great deal of attention...
on mtDNA have produced observations relevant to human evolution.
Age of the common ancestor
By estimating the rate at which mutations occur in mtDNA, the age of the common ancestral mtDNA type can be estimated: "the common ancestral mtDNA (type a) links mtDNA types that have diverged by an average of nearly 0.57%. Assuming a rate of 2%-4% per million years, this implies that the common ancestor of all surviving mtDNA types existed 140,000-290,000 years ago." This observation is robust, and this common direct female line ancestor (or mitochondrial most recent common ancestorMost recent common ancestor
In genetics, the most recent common ancestor of any set of organisms is the most recent individual from which all organisms in the group are directly descended...
(mtMRCA)) of all extant humans has become known as Mitochondrial Eve
Mitochondrial Eve
In the field of human genetics, Mitochondrial Eve refers to the matrilineal "MRCA" . In other words, she was the woman from whom all living humans today descend, on their mother's side, and through the mothers of those mothers and so on, back until all lines converge on one person...
. The observation that the mtMRCA is the direct matrilineal ancestor of all living humans does not mean either that she was the first anatomically modern human, nor that no other female humans lived concurrently with her. Other women would have lived at the same time and passed nuclear genes down to living humans, but their mitochondrial lineages were lost over time. This could be due to random events such as producing only male children.
African origin for modern humans
There is evidence that modern human mtDNA has an African origin: "We infer from the tree of minimum length... that Africa is a likely source of the human mitochondrial gene pool. This inference comes from the observation that one of the two primary branches leads exclusively to African mtDNAs... while the second primary branch also leads to African mtDNAs... By postulating that the common ancestral mtDNA... was African, we minimize the number of intercontinental migrations needed to account for the geographic distribution of mtDNA types."The broad study of African genetic diversity headed by Sarah Tishkoff found the San people
Bushmen
The indigenous people of Southern Africa, whose territory spans most areas of South Africa, Zimbabwe, Lesotho, Mozambique, Swaziland, Botswana, Namibia, and Angola, are variously referred to as Bushmen, San, Sho, Barwa, Kung, or Khwe...
to express the greatest genetic diversity among the 113 distinct populations sampled, making them one of 14 "ancestral population clusters". The research also located the origin of modern human migration in south-western Africa, near the coastal border of Namibia
Namibia
Namibia, officially the Republic of Namibia , is a country in southern Africa whose western border is the Atlantic Ocean. It shares land borders with Angola and Zambia to the north, Botswana to the east and South Africa to the south and east. It gained independence from South Africa on 21 March...
and Angola
Angola
Angola, officially the Republic of Angola , is a country in south-central Africa bordered by Namibia on the south, the Democratic Republic of the Congo on the north, and Zambia on the east; its west coast is on the Atlantic Ocean with Luanda as its capital city...
.
Y chromosome findings
The Y chromosomeY chromosome
The Y chromosome is one of the two sex-determining chromosomes in most mammals, including humans. In mammals, it contains the gene SRY, which triggers testis development if present. The human Y chromosome is composed of about 60 million base pairs...
is much larger than mtDNA, and is relatively homogeneous; therefore it has taken much longer to find distinct lineages and to analyse them. Conversely, because the Y chromosome is so large by comparison, it holds more genetic information. Y chromosome studies show similar findings to those made with mtDNA. The estimate for the age of the ancestral Y chromosome for all extant Y chromosomes is given at about 70,000 years ago and is also placed in Africa; the individual who contributed this Y chromosomal heritage is sometimes referred to as Y chromosome Adam. The difference in dates between Y chromosome Adam and mitochondrial Eve is usually attributed to a higher extinction rate for Y chromosomes due to greater differential reproductive success between individual men, which means that a small number of very successful men may produce many children, while a larger number of less successful men will produce far fewer children.
See also
- The Journey of Man: A Genetic OdysseyThe Journey of Man: A Genetic OdysseyThe Journey of Man: A Genetic Odyssey is a book by Spencer Wells, an American geneticist and anthropologist, in which he uses techniques and theories of genetics and evolutionary biology to trace the geographical dispersal of early human migrations out of Africa...
- Chromosome 2 (human)Chromosome 2 (human)Chromosome 2 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 2 is the second largest human chromosome, spanning more than 237 million base pairs and representing almost 8% of the total DNA in cells.Identifying genes on each...
- Chimpanzee genome projectChimpanzee genome projectThe Chimpanzee Genome Project is an effort to determine the DNA sequence of the Chimpanzee genome. It is expected that by comparing the genomes of humans and other apes, it will be possible to better understand what makes humans distinct from other species....
- Y-DNA haplogroups by ethnic groupsY-DNA haplogroups by ethnic groupsListed here are notable ethnic groups by Y-DNA haplogroups based on relevant studies. The data is presented in two columns for each haplogroup with the first being the sample size and the second the percentage in the haplogroup designated by the column header...
- List of haplogroups of historical and famous figures
- Genetic history of indigenous peoples of the Americas
- Genetic history of the British IslesGenetic history of the British IslesThe genetic history of the British Isles is the subject of research within the larger field of human population genetics. It has developed in parallel with DNA testing technologies capable of identifying genetic similarities and differences between populations...
- Archaeogenetics of the Near EastArchaeogenetics of the Near EastThe archaeogenetics of the Near East involves the study of aDNA or ancient DNA, identifying haplogroups and haplotypes of ancient skeletal remains from both YDNA and mtDNA for populations of the Ancient Near East The archaeogenetics of the Near East involves the study of aDNA or ancient DNA,...
- Genetics and archaeogenetics of South AsiaGenetics and archaeogenetics of South AsiaThe study of the genetics and archaeogenetics of the ethnic groups of South Asia aims at uncovering these groups' genetic history. The geographic position of India makes Indian populations important for the study of the early dispersal of all human populations on the Eurasian continent.The Indian...
- Genetic history of EuropeGenetic history of EuropeThe genetic history of Europe can be inferred from the patterns of genetic diversity across continents and time. The primary data to develop historical scenarios coming from sequences of mitochondrial, Y-chromosome and autosomal DNA from modern populations and if available from ancient DNA...
- Genetic history of ItalyGenetic history of ItalyDuring prehistory Italy was populated by different but very similar Indo-European groups, later collectively listed amongst the Ancient peoples of Italy, of whom the Italic one was predominant....
- HomininaeHomininaeHomininae is a subfamily of Hominidae, which includes humans, gorillas and chimpanzees, and some extinct relatives; it comprises all those hominids, such as Australopithecus, that arose after the split from orangutans . Our family tree, which has 3 main branches leading to chimpanzees, humans and...
- Race and genetics

