
Proteasome
Encyclopedia


Protein complex
A multiprotein complex is a group of two or more associated polypeptide chains. If the different polypeptide chains contain different protein domain, the resulting multiprotein complex can have multiple catalytic functions...
es inside all eukaryote
Eukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
s and archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
, and in some bacteria
Bacteria
Bacteria are a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a wide range of shapes, ranging from spheres to rods and spirals...
. In eukaryotes, they are located in the nucleus
Cell nucleus
In cell biology, the nucleus is a membrane-enclosed organelle found in eukaryotic cells. It contains most of the cell's genetic material, organized as multiple long linear DNA molecules in complex with a large variety of proteins, such as histones, to form chromosomes. The genes within these...
and the cytoplasm
Cytoplasm
The cytoplasm is a small gel-like substance residing between the cell membrane holding all the cell's internal sub-structures , except for the nucleus. All the contents of the cells of prokaryote organisms are contained within the cytoplasm...
. The main function of the proteasome is to degrade unneeded or damaged protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s by proteolysis
Proteolysis
Proteolysis is the directed degradation of proteins by cellular enzymes called proteases or by intramolecular digestion.-Purposes:Proteolysis is used by the cell for several purposes...
, a chemical reaction
Chemical reaction
A chemical reaction is a process that leads to the transformation of one set of chemical substances to another. Chemical reactions can be either spontaneous, requiring no input of energy, or non-spontaneous, typically following the input of some type of energy, such as heat, light or electricity...
that breaks peptide bond
Peptide bond
This article is about the peptide link found within biological molecules, such as proteins. A similar article for synthetic molecules is being created...
s. Enzyme
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
s that carry out such reactions are called protease
Protease
A protease is any enzyme that conducts proteolysis, that is, begins protein catabolism by hydrolysis of the peptide bonds that link amino acids together in the polypeptide chain forming the protein....
s. Proteasomes are part of a major mechanism by which cells
Cell (biology)
The cell is the basic structural and functional unit of all known living organisms. It is the smallest unit of life that is classified as a living thing, and is often called the building block of life. The Alberts text discusses how the "cellular building blocks" move to shape developing embryos....
regulate the concentration
Concentration
In chemistry, concentration is defined as the abundance of a constituent divided by the total volume of a mixture. Four types can be distinguished: mass concentration, molar concentration, number concentration, and volume concentration...
of particular proteins and degrade misfolded proteins
Protein folding
Protein folding is the process by which a protein structure assumes its functional shape or conformation. It is the physical process by which a polypeptide folds into its characteristic and functional three-dimensional structure from random coil....
. The degradation process yields peptide
Peptide
Peptides are short polymers of amino acid monomers linked by peptide bonds. They are distinguished from proteins on the basis of size, typically containing less than 50 monomer units. The shortest peptides are dipeptides, consisting of two amino acids joined by a single peptide bond...
s of about seven to eight amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
s long, which can then be further degraded into amino acids and used in synthesizing
Protein biosynthesis
Protein biosynthesis is the process in which cells build or manufacture proteins. The term is sometimes used to refer only to protein translation but more often it refers to a multi-step process, beginning with amino acid synthesis and transcription of nuclear DNA into messenger RNA, which is then...
new proteins. Proteins are tagged for degradation with a small protein called ubiquitin
Ubiquitin
Ubiquitin is a small regulatory protein that has been found in almost all tissues of eukaryotic organisms. Among other functions, it directs protein recycling.Ubiquitin can be attached to proteins and label them for destruction...
. The tagging reaction is catalyzed by enzymes called ubiquitin ligase
Ubiquitin ligase
A ubiquitin ligase is a protein that in combination with an E2 ubiquitin-conjugating enzyme causes the attachment of ubiquitin to a lysine on a target protein via an isopeptide bond; the E3 ubiquitin ligase targets specific protein substrates for degradation by the proteasome...
s. Once a protein is tagged with a single ubiquitin molecule, this is a signal to other ligases to attach additional ubiquitin molecules. The result is a polyubiquitin chain that is bound by the proteasome, allowing it to degrade the tagged protein.
In structure
Protein structure
Proteins are an important class of biological macromolecules present in all organisms. Proteins are polymers of amino acids. Classified by their physical size, proteins are nanoparticles . Each protein polymer – also known as a polypeptide – consists of a sequence formed from 20 possible L-α-amino...
, the proteasome is a cylindrical complex containing a "core" of four stacked rings around a central pore. Each ring is composed of seven individual proteins. The inner two rings are made of seven β subunits that contain three to seven protease
Protease
A protease is any enzyme that conducts proteolysis, that is, begins protein catabolism by hydrolysis of the peptide bonds that link amino acids together in the polypeptide chain forming the protein....
active site
Active site
In biology the active site is part of an enzyme where substrates bind and undergo a chemical reaction. The majority of enzymes are proteins but RNA enzymes called ribozymes also exist. The active site of an enzyme is usually found in a cleft or pocket that is lined by amino acid residues that...
s. These sites are located on the interior surface of the rings, so that the target protein must enter the central pore before it is degraded. The outer two rings each contain seven α subunits whose function is to maintain a "gate" through which proteins enter the barrel. These α subunits are controlled by binding to "cap" structures or regulatory particles that recognize polyubiquitin tags attached to protein substrates and initiate the degradation process. The overall system of ubiquitination and proteasomal degradation is known as the ubiquitin-proteasome system.
The proteasomal degradation pathway is essential for many cellular processes, including the cell cycle
Cell cycle
The cell cycle, or cell-division cycle, is the series of events that takes place in a cell leading to its division and duplication . In cells without a nucleus , the cell cycle occurs via a process termed binary fission...
, the regulation of gene expression
Gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA , transfer RNA or small nuclear RNA genes, the product is a functional RNA...
, and responses to oxidative stress
Oxidative stress
Oxidative stress represents an imbalance between the production and manifestation of reactive oxygen species and a biological system's ability to readily detoxify the reactive intermediates or to repair the resulting damage...
. The importance of proteolytic degradation inside cells and the role of ubiquitin
Ubiquitin
Ubiquitin is a small regulatory protein that has been found in almost all tissues of eukaryotic organisms. Among other functions, it directs protein recycling.Ubiquitin can be attached to proteins and label them for destruction...
in proteolytic pathways was acknowledged in the award of the 2004 Nobel Prize in Chemistry
Nobel Prize in Chemistry
The Nobel Prize in Chemistry is awarded annually by the Royal Swedish Academy of Sciences to scientists in the various fields of chemistry. It is one of the five Nobel Prizes established by the will of Alfred Nobel in 1895, awarded for outstanding contributions in chemistry, physics, literature,...
to Aaron Ciechanover
Aaron Ciechanover
Aaron Ciechanover is an Israeli biologist, and Nobel laureate in Chemistry.- Biography :Ciechanover was born in Haifa, British mandate of Palestine, a year before the establishment of the State of Israel...
, Avram Hershko
Avram Hershko
Avram Hershko is a Hungarian-Israeli biochemist and Nobel laureate in Chemistry.-Biography:Born Herskó Ferenc in Karcag, Hungary, Hershko emigrated to Israel in 1950. Received his M.D. in 1965 and his Ph.D in 1969 from the Hebrew University-Hadassah Medical School, Jerusalem, Israel...
and Irwin Rose
Irwin Rose
Irwin A. Rose is an American biologist. Along with Aaron Ciechanover and Avram Hershko, he was awarded the 2004 Nobel Prize in Chemistry for the discovery of ubiquitin-mediated protein degradation.-Biography:...
.
Discovery
Before the discovery of the ubiquitin proteasome system, protein degradation in cells was thought to rely mainly on lysosomeLysosome
thumb|350px|Schematic of typical animal cell, showing subcellular components. [[Organelle]]s: [[nucleoli]] [[cell nucleus|nucleus]] [[ribosomes]] [[vesicle |vesicle]] rough [[endoplasmic reticulum]]...
s, membrane-bound organelle
Organelle
In cell biology, an organelle is a specialized subunit within a cell that has a specific function, and is usually separately enclosed within its own lipid bilayer....
s with acid
Acid
An acid is a substance which reacts with a base. Commonly, acids can be identified as tasting sour, reacting with metals such as calcium, and bases like sodium carbonate. Aqueous acids have a pH of less than 7, where an acid of lower pH is typically stronger, and turn blue litmus paper red...
ic and protease
Protease
A protease is any enzyme that conducts proteolysis, that is, begins protein catabolism by hydrolysis of the peptide bonds that link amino acids together in the polypeptide chain forming the protein....
-filled interiors that can degrade and then recycle exogenous proteins and aged or damaged organelles. However, work by Alfred Goldberg in 1977 on ATP-dependent protein degradation in reticulocyte
Reticulocyte
Reticulocytes are immature red blood cells, typically composing about 1% of the red cells in the human body.Reticulocytes develop and mature in the red bone marrow and then circulate for about a day in the blood stream before developing into mature red blood cells. Like mature red blood cells,...
s, which lack lysosomes, suggested the presence of a second intracellular degradation mechanism. This was shown in 1978 to be composed of several distinct protein chains, a novelty among proteases at the time. Later work on modification of histone
Histone
In biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
s led to the identification of an unexpected covalent modification of the histone protein by a bond between a lysine
Lysine
Lysine is an α-amino acid with the chemical formula HO2CCH4NH2. It is an essential amino acid, which means that the human body cannot synthesize it. Its codons are AAA and AAG....
side chain of the histone and the C-terminal glycine
Glycine
Glycine is an organic compound with the formula NH2CH2COOH. Having a hydrogen substituent as its 'side chain', glycine is the smallest of the 20 amino acids commonly found in proteins. Its codons are GGU, GGC, GGA, GGG cf. the genetic code.Glycine is a colourless, sweet-tasting crystalline solid...
residue of ubiquitin
Ubiquitin
Ubiquitin is a small regulatory protein that has been found in almost all tissues of eukaryotic organisms. Among other functions, it directs protein recycling.Ubiquitin can be attached to proteins and label them for destruction...
, a protein that had no known function. It was then discovered that a previously identified protein associated with proteolytic degradation, known as ATP-dependent proteolysis factor 1 (APF-1), was the same protein as ubiquitin. Later, the ATP
Adenosine triphosphate
Adenosine-5'-triphosphate is a multifunctional nucleoside triphosphate used in cells as a coenzyme. It is often called the "molecular unit of currency" of intracellular energy transfer. ATP transports chemical energy within cells for metabolism...
-dependent proteolytic complex that was responsible for ubiquitin-dependent protein degradation was discovered and was called the 26S proteasome.
Much of the early work leading up to the discovery of the ubiquitin proteasome system occurred in the late 1970s and early 1980s at the Technion in the laboratory of Avram Hershko
Avram Hershko
Avram Hershko is a Hungarian-Israeli biochemist and Nobel laureate in Chemistry.-Biography:Born Herskó Ferenc in Karcag, Hungary, Hershko emigrated to Israel in 1950. Received his M.D. in 1965 and his Ph.D in 1969 from the Hebrew University-Hadassah Medical School, Jerusalem, Israel...
, where Aaron Ciechanover
Aaron Ciechanover
Aaron Ciechanover is an Israeli biologist, and Nobel laureate in Chemistry.- Biography :Ciechanover was born in Haifa, British mandate of Palestine, a year before the establishment of the State of Israel...
worked as a graduate student. Hershko's year-long sabbatical in the laboratory of Irwin Rose
Irwin Rose
Irwin A. Rose is an American biologist. Along with Aaron Ciechanover and Avram Hershko, he was awarded the 2004 Nobel Prize in Chemistry for the discovery of ubiquitin-mediated protein degradation.-Biography:...
at the Fox Chase Cancer Center
Fox Chase Cancer Center
The Fox Chase Cancer Center is a National Cancer Institute-designated Comprehensive Cancer Center research facility and hospital located in the Fox Chase section of Philadelphia, Pennsylvania, United States. The main facilities of the center are located on property adjoining Burholme Park...
provided key conceptual insights, though Rose later downplayed his role in the discovery. The three shared the 2004 Nobel Prize in Chemistry
Nobel Prize in Chemistry
The Nobel Prize in Chemistry is awarded annually by the Royal Swedish Academy of Sciences to scientists in the various fields of chemistry. It is one of the five Nobel Prizes established by the will of Alfred Nobel in 1895, awarded for outstanding contributions in chemistry, physics, literature,...
for their work in discovering this system.
Although electron microscopy data revealing the stacked-ring structure of the proteasome became available in the mid-1980s, the first structure of the proteasome core particle was not solved by X-ray crystallography
X-ray crystallography
X-ray crystallography is a method of determining the arrangement of atoms within a crystal, in which a beam of X-rays strikes a crystal and causes the beam of light to spread into many specific directions. From the angles and intensities of these diffracted beams, a crystallographer can produce a...
until 1994. As of 2006, no structure has been solved of the core particle in complex with the most common form of regulatory cap.
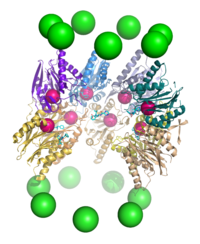
Structure and organization
The proteasome subcomponents are often referred to by their SvedbergSvedberg
A svedberg is a non-SI physical unit used for sedimentation coefficients. It characterizes the behaviour of a particle type in sedimentation processes, notably centrifugation. The svedberg is technically a measure of time, and is defined as exactly 10-13 seconds A svedberg (symbol S, sometimes...
sedimentation coefficient (denoted S). The most common form of the proteasome is known as the 26S proteasome, which is about 2000 kilodaltons
Atomic mass unit
The unified atomic mass unit or dalton is a unit that is used for indicating mass on an atomic or molecular scale. It is defined as one twelfth of the rest mass of an unbound neutral atom of carbon-12 in its nuclear and electronic ground state, and has a value of...
(kDa) in molecular mass
Molecular mass
The molecular mass of a substance is the mass of one molecule of that substance, in unified atomic mass unit u...
and contains one 20S core particle structure and two 19S regulatory caps. The core is hollow and provides an enclosed cavity in which proteins are degraded; openings at the two ends of the core allow the target protein to enter. Each end of the core particle associates with a 19S regulatory subunit that contains multiple ATPase
ATPase
ATPases are a class of enzymes that catalyze the decomposition of adenosine triphosphate into adenosine diphosphate and a free phosphate ion. This dephosphorylation reaction releases energy, which the enzyme harnesses to drive other chemical reactions that would not otherwise occur...
active site
Active site
In biology the active site is part of an enzyme where substrates bind and undergo a chemical reaction. The majority of enzymes are proteins but RNA enzymes called ribozymes also exist. The active site of an enzyme is usually found in a cleft or pocket that is lined by amino acid residues that...
s and ubiquitin binding sites; it is this structure that recognizes polyubiquitinated proteins and transfers them to the catalytic core. An alternative form of regulatory subunit called the 11S particle can associate with the core in essentially the same manner as the 19S particle; the 11S may play a role in degradation of foreign peptides such as those produced after infection by a virus
Virus
A virus is a small infectious agent that can replicate only inside the living cells of organisms. Viruses infect all types of organisms, from animals and plants to bacteria and archaea...
.
20S core particle
The number and diversity of subunits contained in the 20S core particle depends on the organism; the number of distinct and specialized subunits is larger in multicellular than unicellular organisms and larger in eukaryotes than in prokaryotes. All 20S particles consist of four stacked heptameric ring structures that are themselves composed of two different types of subunits; α subunits are structural in nature, whereas β subunits are predominantly catalyticCatalysis
Catalysis is the change in rate of a chemical reaction due to the participation of a substance called a catalyst. Unlike other reagents that participate in the chemical reaction, a catalyst is not consumed by the reaction itself. A catalyst may participate in multiple chemical transformations....
. The outer two rings in the stack consist of seven α subunits each, which serve as docking domains for the regulatory particles and the alpha subunits N-termini form a gate that blocks unregulated access of substrates to the interior cavity. The inner two rings each consist of seven β subunits and contain the protease
Protease
A protease is any enzyme that conducts proteolysis, that is, begins protein catabolism by hydrolysis of the peptide bonds that link amino acids together in the polypeptide chain forming the protein....
active site
Active site
In biology the active site is part of an enzyme where substrates bind and undergo a chemical reaction. The majority of enzymes are proteins but RNA enzymes called ribozymes also exist. The active site of an enzyme is usually found in a cleft or pocket that is lined by amino acid residues that...
s that perform the proteolysis reactions. The size of the proteasome is relatively conserved and is about 150 angstrom
Ångström
The angstrom or ångström, is a unit of length equal to 1/10,000,000,000 of a meter . Its symbol is the Swedish letter Å....
s (Å) by 115 Å. The interior chamber is at most 53 Å wide, though the entrance can be as narrow as 13 Å, suggesting that substrate proteins must be at least partially unfolded to enter.
In archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
such as Thermoplasma acidophilum
Thermoplasma
In taxonomy, Thermoplasma is a genus of the Thermoplasmataceae.Thermoplasma is a genus of archaea. It belongs to the Thermoplasmata, which thrive in acidic and high-temperature environments. Thermoplasma are facultative anaerobes and respire using sulfur and organic carbon...
, all the α and all the β subunits are identical, whereas eukaryotic proteasomes such as those in yeast
Yeast
Yeasts are eukaryotic micro-organisms classified in the kingdom Fungi, with 1,500 species currently described estimated to be only 1% of all fungal species. Most reproduce asexually by mitosis, and many do so by an asymmetric division process called budding...
contain seven distinct types of each subunit. In mammal
Mammal
Mammals are members of a class of air-breathing vertebrate animals characterised by the possession of endothermy, hair, three middle ear bones, and mammary glands functional in mothers with young...
s, the β1, β2, and β5 subunits are catalytic; although they share a common mechanism, they have three distinct substrate specificities considered chymotrypsin
Chymotrypsin
Chymotrypsin is a digestive enzyme that can perform proteolysis. Chymotrypsin preferentially cleaves peptide amide bonds where the carboxyl side of the amide bond is a tyrosine, tryptophan, or phenylalanine. These amino acids contain an aromatic ring in their sidechain that fits into a...
-like, trypsin
Trypsin
Trypsin is a serine protease found in the digestive system of many vertebrates, where it hydrolyses proteins. Trypsin is produced in the pancreas as the inactive proenzyme trypsinogen. Trypsin cleaves peptide chains mainly at the carboxyl side of the amino acids lysine or arginine, except when...
-like, and peptidyl-glutamyl peptide-hydrolyzing (PHGH). Alternative β forms denoted β1i, β2i, and β5i can be expressed in hematopoietic cells in response to exposure to pro-inflammatory
Inflammation
Inflammation is part of the complex biological response of vascular tissues to harmful stimuli, such as pathogens, damaged cells, or irritants. Inflammation is a protective attempt by the organism to remove the injurious stimuli and to initiate the healing process...
signal
Cell signaling
Cell signaling is part of a complex system of communication that governs basic cellular activities and coordinates cell actions. The ability of cells to perceive and correctly respond to their microenvironment is the basis of development, tissue repair, and immunity as well as normal tissue...
s such as cytokine
Cytokine
Cytokines are small cell-signaling protein molecules that are secreted by the glial cells of the nervous system and by numerous cells of the immune system and are a category of signaling molecules used extensively in intercellular communication...
s, in particular, interferon gamma. The proteasome assembled with these alternative subunits is known as the immunoproteasome, whose substrate specificity is altered relative to the normal proteasome.
19S regulatory particle
The 19S particle in eukaryotes consists of 19 individual proteins and is divisible into two subassemblies, a 10-protein base that binds directly to the α ring of the 20S core particle, and a 9-protein lid where polyubiquitin is bound. Six of the ten base proteins are ATPase subunits from the AAA Family, and an evolutionary homolog of these ATPases exists in archaea, called PAN (Proteasome-Activating Nucleotidase). The association of the 19S and 20S particles requires the binding of ATP to the 19S ATPase subunits, and ATP hydrolysis is required for the assembled complex to degrade folded and ubiquitinated proteins. Note that only the step of substrate unfolding requires energy from ATP hydrolysis, while ATP-binding alone can support all the other steps required for protein degradation (e.g., complex assembly, gate opening, translocation, and proteolysis). In fact, ATP binding to the ATPases by itself supports the rapid degradation of unfolded proteins. However, while ATP hydrolysis is required for unfolding only, it is not yet clear whether this energy may be used in the coupling of some of these steps. As of 2011, the atomic structure of the 26S proteasome has not been solved, despite massive efforts to do so. Nevertheless, it is understood, in general, how the 19S associates with and regulates the 20S core particle. In fact, the 19S and 11S particles bind to the same sites in the α rings of the 20S core particle although, they each induce gate opening by different mechanism.Regulation of the 20S by the 19S
The 19S regulatory particle is responsible for stimulating the 20S to degrade proteins. A primary function of the 19S regulatory ATPases is to open the gate in the 20S that blocks the entry of substrates into the degradation chamber. The mechanism by which the proteasomal ATPase open this gate has been recently elucidated. 20S gate opening, and thus substrate degradation, requires the C-termini of the proteasomal ATPases, which contains a specific motifSequence motif
In genetics, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and has, or is conjectured to have, a biological significance...
(i.e., HbYX motif). The ATPases C-termini bind into pockets in the top of the 20S, and tether the ATPase complex to the 20S proteolytic complex, thus joining the substrate unfolding equipment with the 20S degradation machinery. Binding of these C-termini into these 20S pockets by themselves stimulates opening of the gate in the 20S in much the same way that a "key-in-a-lock" opens a door. The precise mechanism by which this "key-in-a-lock" mechanism functions has been structurally elucidated.
11S regulatory particle
20S proteasomes can also associate with a second type of regulatory particle, the 11S regulatory particle, a heptameric structure that does not contain any ATPaseATPase
ATPases are a class of enzymes that catalyze the decomposition of adenosine triphosphate into adenosine diphosphate and a free phosphate ion. This dephosphorylation reaction releases energy, which the enzyme harnesses to drive other chemical reactions that would not otherwise occur...
s and can promote the degradation of short peptide
Peptide
Peptides are short polymers of amino acid monomers linked by peptide bonds. They are distinguished from proteins on the basis of size, typically containing less than 50 monomer units. The shortest peptides are dipeptides, consisting of two amino acids joined by a single peptide bond...
s but not of complete proteins. It is presumed that this is because the complex cannot unfold larger substrates. This structure is also known as PA28 or REG. The mechanisms by which it binds to the core particle through the C-terminal tails of its subunits and induces α-ring conformational change
Conformational change
A macromolecule is usually flexible and dynamic. It can change its shape in response to changes in its environment or other factors; each possible shape is called a conformation, and a transition between them is called a conformational change...
s to open the 20S gate suggest a similar mechanism for the 19S particle. The expression of the 11S particle is induced by interferon gamma and is responsible, in conjunction with the immunoproteasome β subunits, for the generation of peptides that bind to the major histocompatibility complex
Major histocompatibility complex
Major histocompatibility complex is a cell surface molecule encoded by a large gene family in all vertebrates. MHC molecules mediate interactions of leukocytes, also called white blood cells , which are immune cells, with other leukocytes or body cells...
.
Assembly
The assembly of the proteasome is a complex process due to the number of subunits that must associate to form an active complex. The β subunits are synthesized with N-terminal "propeptides" that are post-translationally modified during the assembly of the 20S particle to expose the proteolytic active site. The 20S particle is assembled from two half-proteasomes, each of which consists of a seven-membered pro-β ring attached to a seven-membered α ring. The association of the β rings of the two half-proteasomes triggers threonineThreonine
Threonine is an α-amino acid with the chemical formula HO2CCHCHCH3. Its codons are ACU, ACA, ACC, and ACG. This essential amino acid is classified as polar...
-dependent autolysis of the propeptides to expose the active site. These β interactions are mediated mainly by salt bridges and hydrophobic interactions between conserved alpha helices
Alpha helix
A common motif in the secondary structure of proteins, the alpha helix is a right-handed coiled or spiral conformation, in which every backbone N-H group donates a hydrogen bond to the backbone C=O group of the amino acid four residues earlier...
whose disruption by mutation
Mutation
In molecular biology and genetics, mutations are changes in a genomic sequence: the DNA sequence of a cell's genome or the DNA or RNA sequence of a virus. They can be defined as sudden and spontaneous changes in the cell. Mutations are caused by radiation, viruses, transposons and mutagenic...
damages the proteasome's ability to assemble. The assembly of the half-proteasomes, in turn, is initiated by the assembly of the α subunits into their heptameric ring, forming a template for the association of the corresponding pro-β ring. The assembly of α subunits has not been characterized.
In general, less is known about the assembly and maturation of the 19S regulatory particles. They are believed to assemble as two distinct subcomponents, the ATPase-containing base and the ubiquitin-recognizing lid. The six ATPases in the base may assemble in a pairwise manner mediated by coiled-coil interactions. The order in which the nineteen subunits of the regulatory particle are bound is a likely regulatory mechanism that prevents exposure of the active site before assembly is complete.
The protein degradation process
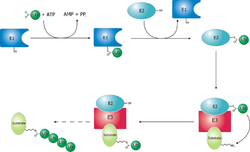
Ubiquitylation and targeting
Proteins are targeted for degradation by the proteasome by covalent modification of a lysineLysine
Lysine is an α-amino acid with the chemical formula HO2CCH4NH2. It is an essential amino acid, which means that the human body cannot synthesize it. Its codons are AAA and AAG....
residue that requires the coordinated reactions of three enzyme
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
s. In the first step, a ubiquitin-activating enzyme
Ubiquitin-activating enzyme
Ubiquitin-activating enzymes, also known as E1 enzymes, catalyze the first step in the ubiquitination reaction, which targets a protein for degradation via a proteasome. This covalent attachment of ubiquitin or ubiquitin-like proteins to targeted proteins is a major mechanism for regulating protein...
(known as E1) hydrolyzes ATP and adenylylates a ubiquitin molecule. This is then transferred to E1's active-site cysteine
Cysteine
Cysteine is an α-amino acid with the chemical formula HO2CCHCH2SH. It is a non-essential amino acid, which means that it is biosynthesized in humans. Its codons are UGU and UGC. The side chain on cysteine is thiol, which is polar and thus cysteine is usually classified as a hydrophilic amino acid...
residue in concert with the adenylylation of a second ubiquitin. This adenylylated ubiquitin is then transferred to a cysteine of a second enzyme, ubiquitin-conjugating enzyme
Ubiquitin-conjugating enzyme
Ubiquitin-conjugating enzymes, also known as E2 enzymes and more rarely as ubiquitin-carrier enzymes, perform the second step in the ubiquitination reaction that targets a protein for degradation via the proteasome.The ubiquitination process covalently attaches ubiquitin, a short protein of 76...
(E2). In the last step, a member of a highly diverse class of enzymes known as ubiquitin ligase
Ubiquitin ligase
A ubiquitin ligase is a protein that in combination with an E2 ubiquitin-conjugating enzyme causes the attachment of ubiquitin to a lysine on a target protein via an isopeptide bond; the E3 ubiquitin ligase targets specific protein substrates for degradation by the proteasome...
s (E3) recognizes the specific protein to be ubiquitinated and catalyzes the transfer of ubiquitin from E2 to this target protein. A target protein must be labeled with at least four ubiquitin monomers (in the form of a polyubiquitin chain) before it is recognized by the proteasome lid. It is therefore the E3 that confers substrate
Substrate (biochemistry)
In biochemistry, a substrate is a molecule upon which an enzyme acts. Enzymes catalyze chemical reactions involving the substrate. In the case of a single substrate, the substrate binds with the enzyme active site, and an enzyme-substrate complex is formed. The substrate is transformed into one or...
specificity to this system. The number of E1, E2, and E3 proteins expressed depends on the organism and cell type, but there are many different E3 enzymes present in humans, indicating that there is a huge number of targets for the ubiquitin proteasome system.
The mechanism by which a polyubiquitinated protein is targeted to the proteasome is not fully understood. Ubiquitin-receptor proteins have an N-terminal
N-terminal end
The N-terminus refers to the start of a protein or polypeptide terminated by an amino acid with a free amine group . The convention for writing peptide sequences is to put the N-terminus on the left and write the sequence from N- to C-terminus...
ubiquitin-like (UBL) domain and one or more ubiquitin-associated (UBA) domains. The UBL domains are recognized by the 19S proteasome caps and the UBA domains bind ubiquitin via three-helix bundles
Helix bundle
A helix bundle is a small protein fold composed of several alpha helices that are usually nearly parallel or antiparallel to each other.-Three-helix bundles:Three-helix bundles are among the smallest and fastest known cooperatively folding structural domains...
. These receptor proteins may escort polyubiquitinated proteins to the proteasome, though the specifics of this interaction and its regulation are unclear.
The ubiquitin protein itself is 76 amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
s long and was named due to its ubiquitous nature, as it has a highly conserved sequence and is found in all known eukaryotic organisms. The genes encoding ubiquitin in eukaryote
Eukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
s are arranged in tandem repeat
Tandem repeat
Tandem repeats occur in DNA when a pattern of two or more nucleotides is repeated and the repetitions are directly adjacent to each other. -Example:An example would be:in which the sequence A-T-T-C-G is repeated three times.-Terminology:...
s, possibly due to the heavy transcription
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
demands on these genes to produce enough ubiquitin for the cell. It has been proposed that ubiquitin is the slowest-evolving
Evolution
Evolution is any change across successive generations in the heritable characteristics of biological populations. Evolutionary processes give rise to diversity at every level of biological organisation, including species, individual organisms and molecules such as DNA and proteins.Life on Earth...
protein identified to date.
Unfolding and translocation
After a protein has been ubiquitinated, it is recognized by the 19S regulatory particle in an ATP-dependent binding step. The substrate protein must then enter the interior of the 20S particle to come in contact with the proteolytic active sites. Because the 20S particle's central channel is narrow and gated by the N-terminal tails of the α ring subunits, the substrates must be at least partially unfolded before they enter the core. The passage of the unfolded substrate into the core is called translocation and necessarily occurs after deubiquitination. However, the order in which substrates are deubiquitinated and unfolded is not yet clear. Which of these processes is the rate-limiting step in the overall proteolysis reaction depends on the specific substrate; for some proteins, the unfolding process is rate-limiting, while deubiquitination is the slowest step for other proteins. The extent to which substrates must be unfolded before translocation is not known, but substantial tertiary structureTertiary structure
In biochemistry and molecular biology, the tertiary structure of a protein or any other macromolecule is its three-dimensional structure, as defined by the atomic coordinates.-Relationship to primary structure:...
, and in particular nonlocal interactions such as disulfide bond
Disulfide bond
In chemistry, a disulfide bond is a covalent bond, usually derived by the coupling of two thiol groups. The linkage is also called an SS-bond or disulfide bridge. The overall connectivity is therefore R-S-S-R. The terminology is widely used in biochemistry...
s, are sufficient to inhibit degradation.
The gate formed by the α subunits prevents peptides longer than about four residues from entering the interior of the 20S particle. The ATP molecules bound before the initial recognition step are hydrolyzed
Hydrolysis
Hydrolysis is a chemical reaction during which molecules of water are split into hydrogen cations and hydroxide anions in the process of a chemical mechanism. It is the type of reaction that is used to break down certain polymers, especially those made by condensation polymerization...
before translocation. While energy is needed for substrate unfolding, it is not required for translocation. The assembled 26S proteasome can degrade unfolded proteins in the presence of a non-hydrolyzable ATP analog, but cannot degrade folded proteins, indicating that energy from ATP hydrolysis is used for substrate unfolding. Passage of the unfolded substrate through the opened gate occurs via facilitated diffusion
Facilitated diffusion
..Facilitated diffusion is a process of passive transport, facilitated by integral proteins. Facilitated diffusion is the spontaneous passage of molecules or ions across a biological membrane passing through specific transmembrane integral proteins...
if the 19S cap is in the ATP-bound state.
The mechanism for unfolding of globular protein
Globular protein
Globular proteins, or spheroproteins are one of the two main protein classes, comprising "globe"-like proteins that are more or less soluble in aqueous solutions...
s is necessarily general, but somewhat dependent on the amino acid sequence
Primary structure
The primary structure of peptides and proteins refers to the linear sequence of its amino acid structural units. The term "primary structure" was first coined by Linderstrøm-Lang in 1951...
. Long sequences of alternating glycine
Glycine
Glycine is an organic compound with the formula NH2CH2COOH. Having a hydrogen substituent as its 'side chain', glycine is the smallest of the 20 amino acids commonly found in proteins. Its codons are GGU, GGC, GGA, GGG cf. the genetic code.Glycine is a colourless, sweet-tasting crystalline solid...
and alanine
Alanine
Alanine is an α-amino acid with the chemical formula CH3CHCOOH. The L-isomer is one of the 20 amino acids encoded by the genetic code. Its codons are GCU, GCC, GCA, and GCG. It is classified as a nonpolar amino acid...
have been shown to inhibit substrate unfolding decreasing the efficiency of proteasomal degradation; this results in the release of partially degraded byproducts, possibly due to the decoupling of the ATP hydrolysis and unfolding steps. Such glycine-alanine repeats are also found in nature, for example in silk
Silk
Silk is a natural protein fiber, some forms of which can be woven into textiles. The best-known type of silk is obtained from the cocoons of the larvae of the mulberry silkworm Bombyx mori reared in captivity...
fibroin
Fibroin
Fibroin is a type of protein created by Bombyx mori in the production of silk. Silk emitted by the silkworm consists of two main proteins, sericin and fibroin, fibroin being the structural center of the silk, and sericin being the sticky material surrounding it.The fibroin protein consists of...
; in particular, certain Epstein-Barr virus
Epstein-Barr virus
The Epstein–Barr virus , also called human herpesvirus 4 , is a virus of the herpes family and is one of the most common viruses in humans. It is best known as the cause of infectious mononucleosis...
gene products bearing this sequence can stall the proteasome, helping the virus propagate by preventing antigen presentation
Antigen presentation
Antigen presentation is a process in the body's immune system by which macrophages, dendritic cells and other cell types capture antigens and then enable their recognition by T-cells....
on the major histocompatibility complex
Major histocompatibility complex
Major histocompatibility complex is a cell surface molecule encoded by a large gene family in all vertebrates. MHC molecules mediate interactions of leukocytes, also called white blood cells , which are immune cells, with other leukocytes or body cells...
.
Proteolysis
The mechanism of proteolysis by the β subunits of the 20S core particle is through a threonineThreonine
Threonine is an α-amino acid with the chemical formula HO2CCHCHCH3. Its codons are ACU, ACA, ACC, and ACG. This essential amino acid is classified as polar...
-dependent nucleophilic attack
Nucleophile
A nucleophile is a species that donates an electron-pair to an electrophile to form a chemical bond in a reaction. All molecules or ions with a free pair of electrons can act as nucleophiles. Because nucleophiles donate electrons, they are by definition Lewis bases.Nucleophilic describes the...
. This mechanism may depend on an associated water
Water
Water is a chemical substance with the chemical formula H2O. A water molecule contains one oxygen and two hydrogen atoms connected by covalent bonds. Water is a liquid at ambient conditions, but it often co-exists on Earth with its solid state, ice, and gaseous state . Water also exists in a...
molecule for deprotonation of the reactive threonine hydroxyl
Hydroxyl
A hydroxyl is a chemical group containing an oxygen atom covalently bonded with a hydrogen atom. In inorganic chemistry, the hydroxyl group is known as the hydroxide ion, and scientists and reference works generally use these different terms though they refer to the same chemical structure in...
. Degradation occurs within the central chamber formed by the association of the two β rings and normally does not release partially degraded products, instead reducing the substrate to short polypeptides typically 7–9 residues long, though they can range from 4 to 25 residues depending on the organism and substrate. The biochemical mechanism that determines product length is not fully characterized. Although the three catalytic β subunits have a common mechanism, they have slightly different substrate specificities, which are considered chymotrypsin
Chymotrypsin
Chymotrypsin is a digestive enzyme that can perform proteolysis. Chymotrypsin preferentially cleaves peptide amide bonds where the carboxyl side of the amide bond is a tyrosine, tryptophan, or phenylalanine. These amino acids contain an aromatic ring in their sidechain that fits into a...
-like, trypsin
Trypsin
Trypsin is a serine protease found in the digestive system of many vertebrates, where it hydrolyses proteins. Trypsin is produced in the pancreas as the inactive proenzyme trypsinogen. Trypsin cleaves peptide chains mainly at the carboxyl side of the amino acids lysine or arginine, except when...
-like, and peptidyl-glutamyl peptide-hydrolyzing (PHGH)-like. These variations in specificity are the result of interatomic contacts with local residues near the active sites of each subunit. Each catalytic β subunit also possesses a conserved lysine
Lysine
Lysine is an α-amino acid with the chemical formula HO2CCH4NH2. It is an essential amino acid, which means that the human body cannot synthesize it. Its codons are AAA and AAG....
residue required for proteolysis.
Although the proteasome normally produces very short peptide fragments, in some cases these products are themselves biologically active and functional molecules. Certain transcription factor
Transcription factor
In molecular biology and genetics, a transcription factor is a protein that binds to specific DNA sequences, thereby controlling the flow of genetic information from DNA to mRNA...
s regulating the expression of specific genes, including one component of the mammal
Mammal
Mammals are members of a class of air-breathing vertebrate animals characterised by the possession of endothermy, hair, three middle ear bones, and mammary glands functional in mothers with young...
ian complex NF-κB, are synthesized as inactive precursors whose ubiquitination and subsequent proteasomal degradation converts them to an active form. Such activity requires the proteasome to cleave the substrate protein internally: rather than processively degrading it from one terminus. It has been suggested that long loops on these proteins' surfaces serve as the proteasomal substrates and enter the central cavity, while the majority of the protein remains outside. Similar effects have been observed in yeast
Yeast
Yeasts are eukaryotic micro-organisms classified in the kingdom Fungi, with 1,500 species currently described estimated to be only 1% of all fungal species. Most reproduce asexually by mitosis, and many do so by an asymmetric division process called budding...
proteins; this mechanism of selective degradation is known as regulated ubiquitin/proteasome dependent processing (RUP).
Ubiquitin-independent degradation
Although most proteasomal substrates must be ubiquitinated before being degraded, there are some exceptions to this general rule, especially when the proteasome plays a normal role in the post-translationalTranslation (genetics)
In molecular biology and genetics, translation is the third stage of protein biosynthesis . In translation, messenger RNA produced by transcription is decoded by the ribosome to produce a specific amino acid chain, or polypeptide, that will later fold into an active protein...
processing of the protein. The proteasomal activation of NF-κB by processing p105 into p50 via internal proteolysis is one major example. Some proteins that are hypothesized to be unstable due to intrinsically unstructured
Intrinsically unstructured proteins
Intrinsically unstructured proteins, often referred to as naturally unfolded proteins or disordered proteins, are proteins characterized by lack of stable tertiary structure when the protein exists as an isolated polypeptide chain under physiological conditions in vitro...
regions, are degraded in a ubiquitin-independent manner. The most well-known example of a ubiquitin-independent proteasome substrate is the enzyme ornithine decarboxylase
Ornithine decarboxylase
The enzyme ornithine decarboxylase catalyzes the decarboxylation of ornithine to form putrescine. This reaction is the committed step in polyamine synthesis. In humans, this protein has 461 amino acids and forms a homodimer....
. Ubiquitin-independent mechanisms targeting key cell cycle
Cell cycle
The cell cycle, or cell-division cycle, is the series of events that takes place in a cell leading to its division and duplication . In cells without a nucleus , the cell cycle occurs via a process termed binary fission...
regulators such as p53
P53
p53 , is a tumor suppressor protein that in humans is encoded by the TP53 gene. p53 is crucial in multicellular organisms, where it regulates the cell cycle and, thus, functions as a tumor suppressor that is involved in preventing cancer...
have also been reported, although p53 is also subject to ubiquitin-dependent degradation. Finally, structurally abnormal, misfolded, or highly oxidized proteins are also subject to ubiquitin-independent and 19S-independent degradation under conditions of cellular stress.
Evolution

Eukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
s. Some prokaryote
Prokaryote
The prokaryotes are a group of organisms that lack a cell nucleus , or any other membrane-bound organelles. The organisms that have a cell nucleus are called eukaryotes. Most prokaryotes are unicellular, but a few such as myxobacteria have multicellular stages in their life cycles...
s, including many archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
and the bacteria
Bacteria
Bacteria are a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a wide range of shapes, ranging from spheres to rods and spirals...
l order Actinomycetales
Actinomycetales
Actinomycetales is an order of Actinobacteria. They are very diverse and contain a variety of subdivisions as well as yet unclassified isolates. This is mainly because some genera are very difficult to classify because of a highly niche-dependent phenotype...
also share homologs of the 20S proteasome, whereas most bacteria possess heat shock
Heat shock
In biochemistry, heat shock is the effect of subjecting a cell to a higher temperature than that of the ideal body temperature of the organism from which the cell line was derived.-Heat shock response:...
genes hslV and hslU, whose gene products are a multimeric protease arranged in a two-layered ring and an ATPase. The hslV protein has been hypothesized to resemble the likely ancestor of the 20S proteasome. In general, HslV is not essential in bacteria, and not all bacteria possess it, whereas some protist
Protist
Protists are a diverse group of eukaryotic microorganisms. Historically, protists were treated as the kingdom Protista, which includes mostly unicellular organisms that do not fit into the other kingdoms, but this group is contested in modern taxonomy...
s possess both the 20S and the hslV systems.
Sequence analysis suggests that the catalytic β subunits diverged earlier in evolution than the predominantly structural α subunits. In bacteria that express a 20S proteasome, the β subunits have high sequence identity to archaeal and eukaryotic β subunits, whereas the α sequence identity is much lower. The presence of 20S proteasomes in bacteria may result from lateral gene transfer, while the diversification of subunits among eukaryotes is ascribed to multiple gene duplication
Gene duplication
Gene duplication is any duplication of a region of DNA that contains a gene; it may occur as an error in homologous recombination, a retrotransposition event, or duplication of an entire chromosome.The second copy of the gene is often free from selective pressure — that is, mutations of it have no...
events.
Cell cycle control
Cell cycle progression is controlled by ordered action of cyclin-dependent kinaseCyclin-dependent kinase
thumb|350px|Schematic of the cell cycle. outer ring: I=[[Interphase]], M=[[Mitosis]]; inner ring: M=Mitosis; G1=[[G1 phase|Gap phase 1]]; S=[[S phase|Synthesis]]; G2=[[G2 phase|Gap phase 2]]...
s (CDKs), activated by specific cyclin
Cyclin
Cyclins are a family of proteins that control the progression of cells through the cell cycle by activating cyclin-dependent kinase enzymes.- Function :...
s that demarcate phases of the cell cycle
Cell cycle
The cell cycle, or cell-division cycle, is the series of events that takes place in a cell leading to its division and duplication . In cells without a nucleus , the cell cycle occurs via a process termed binary fission...
. Mitotic cyclins, which persist in the cell for only a few minutes, have one of the shortest life spans of all intracellular proteins. After a CDK-cyclin complex has performed its function, the associated cyclin is polyubiquitinated and destroyed by the proteasome, which provides directionality for the cell cycle. In particular, exit from mitosis
Mitosis
Mitosis is the process by which a eukaryotic cell separates the chromosomes in its cell nucleus into two identical sets, in two separate nuclei. It is generally followed immediately by cytokinesis, which divides the nuclei, cytoplasm, organelles and cell membrane into two cells containing roughly...
requires the proteasome-dependent dissociation of the regulatory component cyclin B
Cyclin B
Cyclin B is a member of the cyclin family.Cyclin B is a mitotic cyclin. The amount of cyclin B and the activity of the cyclin B-Cdk complex rise through the cell cycle until mitosis, where they fall abruptly due to degradation of cyclin B...
from the mitosis promoting factor complex. In vertebrate
Vertebrate
Vertebrates are animals that are members of the subphylum Vertebrata . Vertebrates are the largest group of chordates, with currently about 58,000 species described. Vertebrates include the jawless fishes, bony fishes, sharks and rays, amphibians, reptiles, mammals, and birds...
cells, "slippage" through the mitotic checkpoint leading to premature M phase exit can occur despite the delay of this exit by the spindle checkpoint
Spindle checkpoint
In order to preserve one cell's identity and its proper functioning, it is necessary to maintain constant the appropriate number of chromosomes after each cell division...
.
Earlier cell cycle checkpoints such as post-restriction point
Restriction point
The restriction point is a G1 phase checkpoint in the cell cycle of animal cells. Prior to the restriction point, a cell exits the cell cycle if specific mitogenic and growth signals are absent. Cells that progress past the restriction point are committed to enter S phase, where DNA synthesis and...
check between G1 phase
G1 phase
The G1 phase is a period in the cell cycle during interphase, before the S phase. For many cells, this phase is the major period of cell growth during its lifespan. During this stage new organelles are being synthesized, so the cell requires both structural proteins and enzymes, resulting in great...
and S phase
S phase
S-phase is the part of the cell cycle in which DNA is replicated, occurring between G1 phase and G2 phase. Precise and accurate DNA replication is necessary to prevent genetic abnormalities which often lead to cell death or disease. Due to the importance, the regulatory pathways that govern this...
similarly involve proteasomal degradation of cyclin A
Cyclin A
Cyclin A is a member of the cyclin family.Cyclin A binds to S phase Cdk2 and is required for the cell to progress through the S phase. Cyclin A/ Cdk2 is inhibited by the complex p21CIP.-External links:*...
, whose ubiquitination is promoted by the anaphase promoting complex (APC), an E3 ubiquitin ligase
Ubiquitin ligase
A ubiquitin ligase is a protein that in combination with an E2 ubiquitin-conjugating enzyme causes the attachment of ubiquitin to a lysine on a target protein via an isopeptide bond; the E3 ubiquitin ligase targets specific protein substrates for degradation by the proteasome...
. The APC and the Skp1/Cul1/F-box protein complex (SCF complex
SCF complex
Skp, Cullin, F-box containing complex is a multi-protein E3 ubiquitin ligase complex catalyzing the ubiquitination of proteins destined for proteasomal degradation...
) are the two key regulators of cyclin degradation and checkpoint control; the SCF itself is regulated by the APC via ubiquitination of the adaptor protein, Skp2, which prevents SCF activity before the G1-S transition.
Individual components of the 19S particle have their own regulatory roles. Gankyrin
Gankyrin
Gankyrin is a recently discovered oncoprotein that is a component of the 19S regulatory cap of the proteasome. Structurally, it contains a 33-amino acid ankyrin repeat that forms a series of alpha helices. It plays a key role in regulating the cell cycle via protein-protein interactions with the...
, a recently identified oncoprotein
Oncogene
An oncogene is a gene that has the potential to cause cancer. In tumor cells, they are often mutated or expressed at high levels.An oncogene is a gene found in the chromosomes of tumor cells whose activation is associated with the initial and continuing conversion of normal cells into cancer...
, is one of the 19S subcomponents that also tightly binds the cyclin-dependent kinase
Cyclin-dependent kinase
thumb|350px|Schematic of the cell cycle. outer ring: I=[[Interphase]], M=[[Mitosis]]; inner ring: M=Mitosis; G1=[[G1 phase|Gap phase 1]]; S=[[S phase|Synthesis]]; G2=[[G2 phase|Gap phase 2]]...
CDK4 and plays a key role in recognizing ubiquitinated p53
P53
p53 , is a tumor suppressor protein that in humans is encoded by the TP53 gene. p53 is crucial in multicellular organisms, where it regulates the cell cycle and, thus, functions as a tumor suppressor that is involved in preventing cancer...
, via its affinity for the ubiquitin ligase MDM2
Mdm2
Mdm2 is an important negative regulator of the p53 tumor suppressor. It is the name of a gene as well as the protein encoded by that gene. Mdm2 protein functions both as an E3 ubiquitin ligase that recognizes the N-terminal trans-activation domain of the p53 tumor suppressor and an inhibitor of...
. Gankyrin is anti-apoptotic
Apoptosis
Apoptosis is the process of programmed cell death that may occur in multicellular organisms. Biochemical events lead to characteristic cell changes and death. These changes include blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, and chromosomal DNA fragmentation...
and has been shown to be overexpressed in some tumor
Tumor
A tumor or tumour is commonly used as a synonym for a neoplasm that appears enlarged in size. Tumor is not synonymous with cancer...
cell types such as hepatocellular carcinoma
Hepatocellular carcinoma
Hepatocellular carcinoma is the most common type of liver cancer. Most cases of HCC are secondary to either a viral hepatitide infection or cirrhosis .Compared to other cancers, HCC is quite a rare tumor in the United States...
.
Regulation of plant growth
In plantPlant
Plants are living organisms belonging to the kingdom Plantae. Precise definitions of the kingdom vary, but as the term is used here, plants include familiar organisms such as trees, flowers, herbs, bushes, grasses, vines, ferns, mosses, and green algae. The group is also called green plants or...
s, signaling by auxin
Auxin
Auxins are a class of plant hormones with some morphogen-like characteristics. Auxins have a cardinal role in coordination of many growth and behavioral processes in the plant's life cycle and are essential for plant body development. Auxins and their role in plant growth were first described by...
s, or phytohormones that order the direction and tropism
Tropism
A tropism is a biological phenomenon, indicating growth or turning movement of a biological organism, usually a plant, in response to an environmental stimulus. In tropisms, this response is dependent on the direction of the stimulus...
of plant growth, induces the targeting of a class of transcription factor
Transcription factor
In molecular biology and genetics, a transcription factor is a protein that binds to specific DNA sequences, thereby controlling the flow of genetic information from DNA to mRNA...
repressors known as Aux/IAA proteins for proteasomal degradation. These proteins are ubiquitinated by SCFTIR1, or SCF in complex with the auxin receptor TIR1. Degradation of Aux/IAA proteins derepresses transcription factors in the auxin-response factor (ARF) family and induces ARF-directed gene expression. The cellular consequences of ARF activation depend on the plant type and developmental stage, but are involved in directing growth in roots and leaf veins. The specific response to ARF derepression is thought to be mediated by specificity in the pairing of individual ARF and Aux/IAA proteins.
Apoptosis
Both internal and external signalsCell signaling
Cell signaling is part of a complex system of communication that governs basic cellular activities and coordinates cell actions. The ability of cells to perceive and correctly respond to their microenvironment is the basis of development, tissue repair, and immunity as well as normal tissue...
can lead to the induction of apoptosis
Apoptosis
Apoptosis is the process of programmed cell death that may occur in multicellular organisms. Biochemical events lead to characteristic cell changes and death. These changes include blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, and chromosomal DNA fragmentation...
, or programmed cell death. The resulting deconstruction of cellular components is primarily carried out by specialized proteases known as caspase
Caspase
Caspases, or cysteine-aspartic proteases or cysteine-dependent aspartate-directed proteases are a family of cysteine proteases that play essential roles in apoptosis , necrosis, and inflammation....
s, but the proteasome also plays important and diverse roles in the apoptotic process. The involvement of the proteasome in this process is indicated by both the increase in protein ubiquitination, and of E1, E2, and E3 enzymes that is observed well in advance of apoptosis, During apoptosis, proteasomes localized to the nucleus have also been observed to translocate to outer membrane blebs characteristic of apoptosis.
Proteasome inhibition has different effects on apoptosis induction in different cell types. In general, the proteasome is not required for apoptosis, although inhibiting it is pro-apoptotic in most cell types that have been studied. Apoptosis is mediated through disrupting the regulated degradation of pro-growth cell cycle proteins. However, some cell lines — in particular, primary cultures of quiescent
G0 phase
The G0 phase is a period in the cell cycle in which cells exist in a quiescent state. G0 phase is viewed as either an extended G1 phase, where the cell is neither dividing nor preparing to divide, or a distinct quiescent stage that occurs outside of the cell cycle...
and differentiated cells such as thymocyte
Thymocyte
Thymocytes are hematopoietic progenitor cells present in the thymus. Thymopoiesis is the process in the thymus by which thymocytes differentiate into mature T lymphocytes. The primary function of thymocytes is the generation of T lymphocytes . The thymus provides an inductive environment, which...
s and neuron
Neuron
A neuron is an electrically excitable cell that processes and transmits information by electrical and chemical signaling. Chemical signaling occurs via synapses, specialized connections with other cells. Neurons connect to each other to form networks. Neurons are the core components of the nervous...
s — are prevented from undergoing apoptosis on exposure to proteasome inhibitors. The mechanism for this effect is not clear, but is hypothesized to be specific to cells in quiescent states, or to result from the differential activity of the pro-apoptotic kinase
Kinase
In chemistry and biochemistry, a kinase is a type of enzyme that transfers phosphate groups from high-energy donor molecules, such as ATP, to specific substrates, a process referred to as phosphorylation. Kinases are part of the larger family of phosphotransferases...
JNK. The ability of proteasome inhibitors to induce apoptosis in rapidly dividing cells has been exploited in several recently developed chemotherapy
Chemotherapy
Chemotherapy is the treatment of cancer with an antineoplastic drug or with a combination of such drugs into a standardized treatment regimen....
agents such as bortezomib
Bortezomib
Bortezomib is the first therapeutic proteasome inhibitor to be tested in humans. It is approved in the U.S. for treating relapsed multiple myeloma and mantle cell lymphoma...
and .
Response to cellular stress
In response to cellular stresses – such as infectionInfection
An infection is the colonization of a host organism by parasite species. Infecting parasites seek to use the host's resources to reproduce, often resulting in disease...
, heat shock
Heat shock
In biochemistry, heat shock is the effect of subjecting a cell to a higher temperature than that of the ideal body temperature of the organism from which the cell line was derived.-Heat shock response:...
, or oxidative damage
Reactive oxygen species
Reactive oxygen species are chemically reactive molecules containing oxygen. Examples include oxygen ions and peroxides. Reactive oxygen species are highly reactive due to the presence of unpaired valence shell electrons....
– heat shock protein
Heat shock protein
Heat shock proteins are a class of functionally related proteins involved in the folding and unfolding of other proteins. Their expression is increased when cells are exposed to elevated temperatures or other stress. This increase in expression is transcriptionally regulated...
s that identify misfolded or unfolded proteins and target them for proteasomal degradation are expressed. Both Hsp27
Hsp27
Heat shock protein 27 also known as heat shock protein beta-1 is a protein that in humans is encoded by the HSPB1 gene.Hsp27 is a chaperone of the sHsp group among ubiquitin, α-crystallin, Hsp20 and others...
and Hsp90
Hsp90
Hsp90 is a molecular chaperone and is one of the most abundant proteins expressed in cells. It is a member of the heat shock protein family, which is upregulated in response to stress...
—chaperone proteins have been implicated in increasing the activity of the ubiquitin-proteasome system, though they are not direct participants in the process. Hsp70
Hsp70
The 70 kilodalton heat shock proteins are a family of ubiquitously expressed heat shock proteins. Proteins with similar structure exist in virtually all living organisms...
, on the other hand, binds exposed hydrophobic patches on the surface of misfolded proteins and recruits E3 ubiquitin ligases such as CHIP to tag the proteins for proteasomal degradation. The CHIP protein (carboxyl terminus of Hsp70-interacting protein) is itself regulated via inhibition of interactions between the E3 enzyme CHIP and its E2 binding partner.
Similar mechanisms exist to promote the degradation of oxidatively damaged
Redox
Redox reactions describe all chemical reactions in which atoms have their oxidation state changed....
proteins via the proteasome system. In particular, proteasomes localized to the nucleus are regulated by PARP and actively degrade inappropriately oxidized histone
Histone
In biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation...
s. Oxidized proteins, which often form large amorphous aggregates in the cell, can be degraded directly by the 20S core particle without the 19S regulatory cap and do not require ATP hydrolysis or tagging with ubiquitin. However, high levels of oxidative damage increases the degree of cross-linking between protein fragments, rendering the aggregates resistant to proteolysis. Larger numbers and sizes of such highly oxidized aggregates are associated with aging.
Dysregulation of the ubiquitin proteasome system may contribute to several neural diseases. It may lead to brain tumors such as astrocytomas. In some of the late-onset neurodegenerative diseases that share aggregation of misfolded proteins as a common feature, such as Parkinson's disease
Parkinson's disease
Parkinson's disease is a degenerative disorder of the central nervous system...
and Alzheimer's disease
Alzheimer's disease
Alzheimer's disease also known in medical literature as Alzheimer disease is the most common form of dementia. There is no cure for the disease, which worsens as it progresses, and eventually leads to death...
, large insoluble aggregates of misfolded proteins can form and then result in neurotoxicity
Neurotoxicity
Neurotoxicity occurs when the exposure to natural or artificial toxic substances, which are called neurotoxins, alters the normal activity of the nervous system in such a way as to cause damage to nervous tissue. This can eventually disrupt or even kill neurons, key cells that transmit and process...
, through mechanisms that are not yet well understood. Decreased proteasome activity has been suggested as a cause of aggregation and Lewy body
Lewy body
Lewy bodies are abnormal aggregates of protein that develop inside nerve cells in Parkinson's disease , Lewy Body Dementia and some other disorders. They are identified under the microscope when histology is performed on the brain....
formation in Parkinson's. This hypothesis is supported by the observation that yeast
Yeast
Yeasts are eukaryotic micro-organisms classified in the kingdom Fungi, with 1,500 species currently described estimated to be only 1% of all fungal species. Most reproduce asexually by mitosis, and many do so by an asymmetric division process called budding...
models of Parkinson's are more susceptible to toxicity from α-synuclein
Alpha-synuclein
Alpha-synuclein is a protein that, in humans, is encoded by the SNCA gene. An alpha-synuclein fragment, known as the non-Abeta component of Alzheimer's disease amyloid, originally found in an amyloid-enriched fraction, is shown to be a fragment of its precursor protein, NACP, by cloning of the...
, the major protein component of Lewy bodies, under conditions of low proteasome activity. Impaired proteasomal activity may underlie cognitive disorders such as the autism spectrum disorders, and muscle and nerve diseases such as inclusion body myopathy.
Role in the immune system
The proteasome plays a straightforward but critical role in the function of the adaptive immune system. Peptide antigenAntigen
An antigen is a foreign molecule that, when introduced into the body, triggers the production of an antibody by the immune system. The immune system will then kill or neutralize the antigen that is recognized as a foreign and potentially harmful invader. These invaders can be molecules such as...
s are displayed by the major histocompatibility complex
Major histocompatibility complex
Major histocompatibility complex is a cell surface molecule encoded by a large gene family in all vertebrates. MHC molecules mediate interactions of leukocytes, also called white blood cells , which are immune cells, with other leukocytes or body cells...
class I (MHC) proteins on the surface of antigen-presenting cell
Antigen-presenting cell
An antigen-presenting cell or accessory cell is a cell that displays foreign antigen complexes with major histocompatibility complex on their surfaces. T-cells may recognize these complexes using their T-cell receptors...
s. These peptides are products of proteasomal degradation of proteins originated by the invading pathogen
Pathogen
A pathogen gignomai "I give birth to") or infectious agent — colloquially, a germ — is a microbe or microorganism such as a virus, bacterium, prion, or fungus that causes disease in its animal or plant host...
. Although constitutively expressed proteasomes can participate in this process, a specialized complex composed of proteins whose expression
Gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA , transfer RNA or small nuclear RNA genes, the product is a functional RNA...
is induced by interferon gamma produces peptides of the optimal size and composition for MHC binding. These proteins whose expression increases during the immune response include the 11S regulatory particle, whose main known biological role is regulating the production of MHC ligands, and specialized β subunits called β1i, β2i, and β5i with altered substrate specificity. The complex formed with the specialized β subunits is known as the immunoproteasome. Another β5i variant subunit, β5t, is expressed in the thymus, leading to a thymus-specific "thymoproteasome" whose function is as yet unclear.
The strength of MHC class I ligand binding is dependent on the composition of the ligand C-terminus
C-terminal end
The C-terminus is the end of an amino acid chain , terminated by a free carboxyl group . When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus...
, as peptides bind by hydrogen bond
Hydrogen bond
A hydrogen bond is the attractive interaction of a hydrogen atom with an electronegative atom, such as nitrogen, oxygen or fluorine, that comes from another molecule or chemical group. The hydrogen must be covalently bonded to another electronegative atom to create the bond...
ing and by close contacts with a region called the "B pocket" on the MHC surface. Many MHC class I alleles prefer hydrophobic C-terminal residues, and the immunoproteasome complex is more likely to generate hydrophobic C-termini.
Due to its role in generating the activated form of NF-κB, an anti-apoptotic
Apoptosis
Apoptosis is the process of programmed cell death that may occur in multicellular organisms. Biochemical events lead to characteristic cell changes and death. These changes include blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, and chromosomal DNA fragmentation...
and pro-inflammatory
Inflammation
Inflammation is part of the complex biological response of vascular tissues to harmful stimuli, such as pathogens, damaged cells, or irritants. Inflammation is a protective attempt by the organism to remove the injurious stimuli and to initiate the healing process...
regulator of cytokine
Cytokine
Cytokines are small cell-signaling protein molecules that are secreted by the glial cells of the nervous system and by numerous cells of the immune system and are a category of signaling molecules used extensively in intercellular communication...
expression, proteasomal activity has been linked to inflammatory and autoimmune disease
Autoimmune disease
Autoimmune diseases arise from an overactive immune response of the body against substances and tissues normally present in the body. In other words, the body actually attacks its own cells. The immune system mistakes some part of the body as a pathogen and attacks it. This may be restricted to...
s. Increased levels of proteasome activity correlate with disease activity and have been implicated in autoimmune diseases including systemic lupus erythematosus
Systemic lupus erythematosus
Systemic lupus erythematosus , often abbreviated to SLE or lupus, is a systemic autoimmune disease that can affect any part of the body. As occurs in other autoimmune diseases, the immune system attacks the body's cells and tissue, resulting in inflammation and tissue damage...
and rheumatoid arthritis
Rheumatoid arthritis
Rheumatoid arthritis is a chronic, systemic inflammatory disorder that may affect many tissues and organs, but principally attacks synovial joints. The process produces an inflammatory response of the synovium secondary to hyperplasia of synovial cells, excess synovial fluid, and the development...
.
The proteasome is also involved in Intracellular antibody-mediated proteolysis of antibody bound virions. In this neutralisation pathway, TRIM21
TRIM21
Tripartite motif-containing protein 21 also known as E3 ubiquitin-protein ligase TRIM21 is a protein that in humans is encoded by the TRIM21 gene. Alternatively spliced transcript variants for this gene have been described but the full-length nature of only one has been determined...
(a protein of the tripartite motif family) binds with immunoglobulin G
Immunoglobulin G
Immunoglobulin G are antibody molecules. Each IgG is composed of four peptide chains — two heavy chains γ and two light chains. Each IgG has two antigen binding sites. Other immunoglobulins may be described in terms of polymers with the IgG structure considered the monomer.IgG constitutes 75%...
to direct the virion to the proteasome where it is degraded.
Proteasome inhibitors
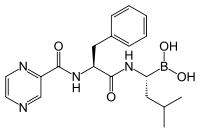
Proteasome inhibitor
Proteasome inhibitors are drugs that block the action of proteasomes, cellular complexes that break down proteins, like the p53 protein. Proteasome inhibitors are being studied in the treatment of cancer.-Examples:...
s have effective anti-tumor
Tumor
A tumor or tumour is commonly used as a synonym for a neoplasm that appears enlarged in size. Tumor is not synonymous with cancer...
activity in cell culture
Cell culture
Cell culture is the complex process by which cells are grown under controlled conditions. In practice, the term "cell culture" has come to refer to the culturing of cells derived from singlecellular eukaryotes, especially animal cells. However, there are also cultures of plants, fungi and microbes,...
, inducing apoptosis
Apoptosis
Apoptosis is the process of programmed cell death that may occur in multicellular organisms. Biochemical events lead to characteristic cell changes and death. These changes include blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, and chromosomal DNA fragmentation...
by disrupting the regulated degradation of pro-growth cell cycle proteins. This approach of selectively inducing apoptosis in tumor cells has proven effective in animal models and human trials. Bortezomib
Bortezomib
Bortezomib is the first therapeutic proteasome inhibitor to be tested in humans. It is approved in the U.S. for treating relapsed multiple myeloma and mantle cell lymphoma...
, a molecule developed by Millennium Pharmaceuticals
Millennium Pharmaceuticals
Millennium Pharmaceuticals, Inc., The Takeda Oncology Company is a biopharmaceutical company based in Cambridge, Massachusetts. The Company markets Velcade for injection, a cancer product, and has a growing clinical development pipeline of product candidates...
and marketed as Velcade, is the first proteasome inhibitor to reach clinical use as a chemotherapy
Chemotherapy
Chemotherapy is the treatment of cancer with an antineoplastic drug or with a combination of such drugs into a standardized treatment regimen....
agent. Bortezomib is used in the treatment of multiple myeloma
Multiple myeloma
Multiple myeloma , also known as plasma cell myeloma or Kahler's disease , is a cancer of plasma cells, a type of white blood cell normally responsible for the production of antibodies...
. Notably, multiple myeloma has been observed to result in increased proteasome levels in blood serum
Blood serum
In blood, the serum is the component that is neither a blood cell nor a clotting factor; it is the blood plasma with the fibrinogens removed...
that decrease to normal levels in response to successful chemotherapy. Studies in animals have indicated that bortezomib may also have clinically significant effects in pancreatic cancer
Pancreatic cancer
Pancreatic cancer refers to a malignant neoplasm of the pancreas. The most common type of pancreatic cancer, accounting for 95% of these tumors is adenocarcinoma, which arises within the exocrine component of the pancreas. A minority arises from the islet cells and is classified as a...
. Preclinical and early clinical studies have been started to examine bortezomib's effectiveness in treating other B-cell-related cancers, particularly some types of non-Hodgkin's lymphoma.
The molecule ritonavir
Ritonavir
Ritonavir, with trade name Norvir , is an antiretroviral drug from the protease inhibitor class used to treat HIV infection and AIDS....
, marketed as Norvir, was developed as a protease inhibitor
Protease inhibitor (pharmacology)
Protease inhibitors are a class of drugs used to treat or prevent infection by viruses, including HIV and Hepatitis C. PIs prevent viral replication by inhibiting the activity of proteases, e.g.HIV-1 protease, enzymes used by the viruses to cleave nascent proteins for final assembly of new...
and used to target HIV
HIV
Human immunodeficiency virus is a lentivirus that causes acquired immunodeficiency syndrome , a condition in humans in which progressive failure of the immune system allows life-threatening opportunistic infections and cancers to thrive...
infection. However, it has been shown to inhibit proteasomes as well as free proteases; to be specific, the chymotrypsin
Chymotrypsin
Chymotrypsin is a digestive enzyme that can perform proteolysis. Chymotrypsin preferentially cleaves peptide amide bonds where the carboxyl side of the amide bond is a tyrosine, tryptophan, or phenylalanine. These amino acids contain an aromatic ring in their sidechain that fits into a...
-like activity of the proteasome is inhibited by ritonavir, while the trypsin
Trypsin
Trypsin is a serine protease found in the digestive system of many vertebrates, where it hydrolyses proteins. Trypsin is produced in the pancreas as the inactive proenzyme trypsinogen. Trypsin cleaves peptide chains mainly at the carboxyl side of the amino acids lysine or arginine, except when...
-like activity is somewhat enhanced. Studies in animal models suggest that ritonavir may have inhibitory effects on the growth of glioma
Glioma
A glioma is a type of tumor that starts in the brain or spine. It is called a glioma because it arises from glial cells. The most common site of gliomas is the brain.-By type of cell:...
cells.
Proteasome inhibitors have also shown promise in treating autoimmune diseases in animal models. For example, studies in mice bearing human skin grafts found a reduction in the size of lesions from psoriasis
Psoriasis
Psoriasis is an autoimmune disease that appears on the skin. It occurs when the immune system mistakes the skin cells as a pathogen, and sends out faulty signals that speed up the growth cycle of skin cells. Psoriasis is not contagious. However, psoriasis has been linked to an increased risk of...
after treatment with a proteasome inhibitor. Inhibitors also show positive effects in rodent
Rodent
Rodentia is an order of mammals also known as rodents, characterised by two continuously growing incisors in the upper and lower jaws which must be kept short by gnawing....
models of asthma
Asthma
Asthma is the common chronic inflammatory disease of the airways characterized by variable and recurring symptoms, reversible airflow obstruction, and bronchospasm. Symptoms include wheezing, coughing, chest tightness, and shortness of breath...
.
Labeling and inhibition of the proteasome is also of interest in laboratory settings for both in vitro and in vivo study of proteasomal activity in cells. The most commonly used laboratory inhibitors are lactacystin
Lactacystin
Lactacystin is an organic compound naturally synthesized by bacteria of the genus Streptomyces first described in 1991. The first total synthesis of lactacystin was developed by Elias Corey in 1992. The molecule is most commonly used as in biochemistry and cell biology laboratories as a selective...
, a natural product synthesized by Streptomyces
Streptomyces
Streptomyces is the largest genus of Actinobacteria and the type genus of the family Streptomycetaceae. Over 500 species of Streptomyces bacteria have been described. As with the other Actinobacteria, streptomycetes are gram-positive, and have genomes with high guanine and cytosine content...
bacteria
Bacteria
Bacteria are a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a wide range of shapes, ranging from spheres to rods and spirals...
, and peptide MG132
MG132
MG132 is a specific, potent, reversible, and cell-permeable proteasome inhibitor . Reduces the degradation of ubiquitin-conjugated proteins in mammalian cells and permeable strains of yeast by the 26S complex without affecting its ATPase or isopeptidase activities. MG132 activates c-Jun N-terminal...
. Fluorescent inhibitors have also been developed to specifically label the active sites of the assembled proteasome.
See also
- The Proteolysis MapThe Proteolysis MapThe Proteolysis MAP is an integrated web resource focused on proteases.-Rationale:PMAP is to aid the protease researchers in reasoning about proteolytic networks and metabolic pathways.-History and funding:...
- ExosomeExosome complexThe exosome complex is a multi-protein complex capable of degrading various types of RNA molecules...
- Endoplasmic reticulum-associated protein degradationERADEndoplasmic-reticulum-associated protein degradation designates a cellular pathway which targets misfolded proteins of the endoplasmic reticulum for ubiquitination and subsequent degradation by a protein-degrading complex, called the proteasome....

